9 0 5 7 8
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 2.6 Breast Cancer (Dove Med Press)
- 3.9 Clin Epidemiol
- 3.3 Cancer Manag Res
- 3.9 Infect Drug Resist
- 3.6 Clin Interv Aging
- 4.8 Drug Des Dev Ther
- 2.8 Int J Chronic Obstr
- 8.0 Int J Nanomed
- 2.3 Int J Women's Health
- 3.2 Neuropsych Dis Treat
- 4.0 OncoTargets Ther
- 2.2 Patient Prefer Adher
- 2.8 Ther Clin Risk Manag
- 2.7 J Pain Res
- 3.3 Diabet Metab Synd Ob
- 4.3 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.9 Pharmgenomics Pers Med
- 3.5 Risk Manag Healthc Policy
- 4.5 J Inflamm Res
- 2.3 Int J Gen Med
- 4.1 J Hepatocell Carcinoma
- 3.2 J Asthma Allergy
- 2.3 Clin Cosmet Investig Dermatol
- 3.3 J Multidiscip Healthc

基于 WGCNA 确定乳腺癌的重要模块和生物标志物
Authors Tian Z, He W, Tang J, Liao X, Yang Q, Wu Y, Wu G
Received 16 April 2020
Accepted for publication 17 June 2020
Published 12 July 2020 Volume 2020:13 Pages 6805—6817
DOI https://doi.org/10.2147/OTT.S258439
Checked for plagiarism Yes
Review by Single-blind
Peer reviewer comments 2
Editor who approved publication: Dr Cho
Introduction: Breast cancer (BRCA) has the highest incidence among female malignancies, and the prognosis for these patients remains poor.
Materials and Methods: In this study, core modules and central genes related to BRCA were identified through a weighted gene co-expression network analysis (WGCNA). Gene expression profiles and clinical data of GSE25066 were obtained from the Gene Expression Omnibus (GEO) database. The result was validated with RNA-seq data from The Cancer Genome Atlas (TCGA) and Oncomine database. The top 30 key module genes with the highest intramodule connectivity were selected as the core genes (R2 = 0.40).
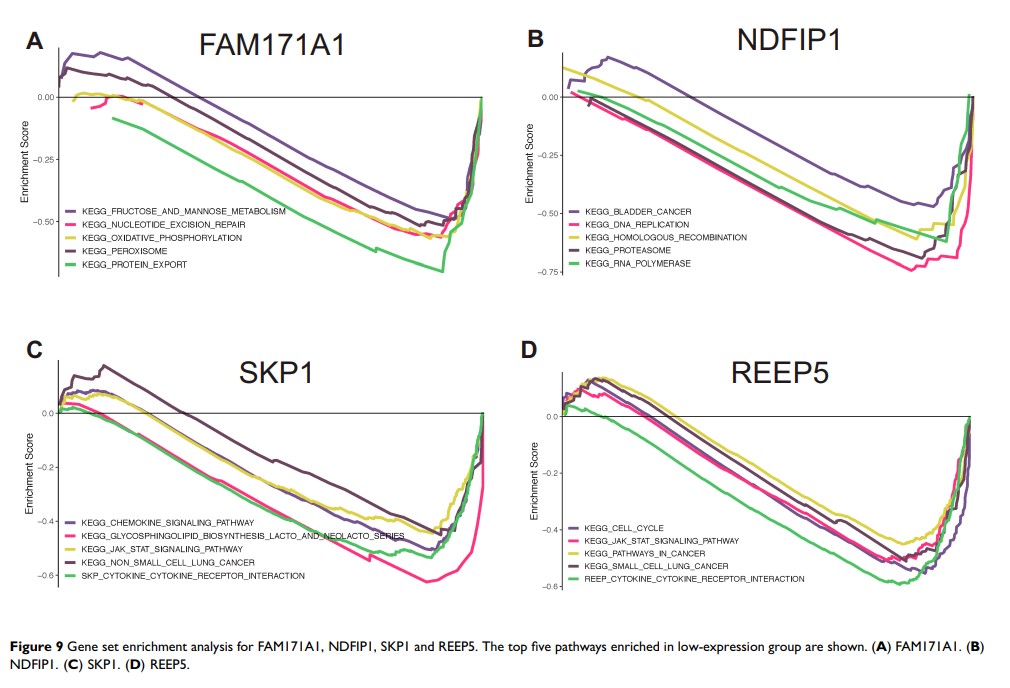
Results: According to TCGA and Oncomine datasets, seven genes were selected as candidate hub genes. Following further experimental verification, four hub genes (FAM171A1, NDFIP1, SKP1, and REEP5) were retained.
Conclusion: We identified four hub genes as candidate biomarkers for BRCA. These hub genes may provide a theoretical basis for targeted therapy against BRCA.
Keywords: breast cancer, WGCNA, GEO, Oncomine, prognosis