9 0 5 7 8
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 2.6 Breast Cancer (Dove Med Press)
- 3.9 Clin Epidemiol
- 3.3 Cancer Manag Res
- 3.9 Infect Drug Resist
- 3.6 Clin Interv Aging
- 4.8 Drug Des Dev Ther
- 2.8 Int J Chronic Obstr
- 8.0 Int J Nanomed
- 2.3 Int J Women's Health
- 3.2 Neuropsych Dis Treat
- 4.0 OncoTargets Ther
- 2.2 Patient Prefer Adher
- 2.8 Ther Clin Risk Manag
- 2.7 J Pain Res
- 3.3 Diabet Metab Synd Ob
- 4.3 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.9 Pharmgenomics Pers Med
- 3.5 Risk Manag Healthc Policy
- 4.5 J Inflamm Res
- 2.3 Int J Gen Med
- 4.1 J Hepatocell Carcinoma
- 3.2 J Asthma Allergy
- 2.3 Clin Cosmet Investig Dermatol
- 3.3 J Multidiscip Healthc

乙型肝炎病毒相关性肝细胞癌中切除修复交叉互补酶 mRNA 表达的诊断和预后价值
Authors Yang L, Xu M, Cui CB, Wei PH, Wu SZ, Cen ZJ, Meng XX, Huang QG, Xie ZC
Received 3 July 2018
Accepted for publication 7 September 2018
Published 5 November 2018 Volume 2018:10 Pages 5313—5328
DOI https://doi.org/10.2147/CMAR.S179043
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Colin Mak
Peer reviewer comments 4
Editor who approved publication: Dr Antonella D'Anneo
Background: The current study aims at using the whole genome expression
profile chips for systematically investigating the diagnostic and prognostic
values of excision repair cross-complementation (ERCC) genes in hepatitis B
virus (HBV)-related hepatocellular carcinoma (HCC).
Materials and
methods: Whole genome expression profile
chips were obtained from the GSE14520. The receiver-operating characteristic
(ROC) curve, survival analysis, and nomogram were used to investigate the
diagnostic and prognostic values of ERCC genes. Investigation of the potential
function of ERCC8 was
carried out by gene set enrichment analysis (GSEA) and genome-wide coexpression
analysis.
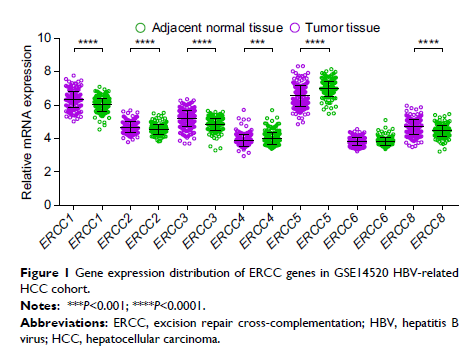
Results: ROC analysis suggests that six ERCC genes (ERCC1 , ERCC2 , ERCC3 , ERCC4 , ERCC5 , and ERCC8 ) were dysregulated and may
have potential to distinguish between HBV-related HCC tumor and paracancerous
tissues (area under the curve of ROC ranged from 0.623 to 0.744). Survival
analysis demonstrated that high ERCC8 expression
was associated with a significantly decreased risk of recurrence
(adjusted P =0.021; HR=0.643; 95%
CI=0.442–0.937) and death (adjusted P =0.049; HR=0.631;
95% CI=0.399–0.998) in HBV-related HCC. Then, we also developed two nomograms
for the HBV-related HCC individualized prognosis predictions. GSEA suggests
that the high expression of ERCC8 may have
involvement in the energy metabolism biological processes. As the genome-wide
coexpression analysis and functional assessment of ERCC8 suggest, those
coexpressed genes were significantly enriched in multiple biological processes
of DNA damage and repair.
Conclusion: The present study indicates that six ERCC genes (ERCC1 , ERCC2 , ERCC3 , ERCC4 , ERCC5 , and ERCC8 ) were dysregulated
between HBV-related HCC tumor and paracancerous tissues and that the mRNA
expression of ERCC8 may
serve as a potential biomarker for the HBV-related HCC prognosis.
Keywords: HBV, hepatocellular carcinoma, diagnosis, prognosis, ERCC