9 0 5 7 8
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 2.6 Breast Cancer (Dove Med Press)
- 3.9 Clin Epidemiol
- 3.3 Cancer Manag Res
- 3.9 Infect Drug Resist
- 3.6 Clin Interv Aging
- 4.8 Drug Des Dev Ther
- 2.8 Int J Chronic Obstr
- 8.0 Int J Nanomed
- 2.3 Int J Women's Health
- 3.2 Neuropsych Dis Treat
- 4.0 OncoTargets Ther
- 2.2 Patient Prefer Adher
- 2.8 Ther Clin Risk Manag
- 2.7 J Pain Res
- 3.3 Diabet Metab Synd Ob
- 4.3 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.9 Pharmgenomics Pers Med
- 3.5 Risk Manag Healthc Policy
- 4.5 J Inflamm Res
- 2.3 Int J Gen Med
- 4.1 J Hepatocell Carcinoma
- 3.2 J Asthma Allergy
- 2.3 Clin Cosmet Investig Dermatol
- 3.3 J Multidiscip Healthc

已发表论文
miRNA 相关基因的单核苷酸多态性和在非霍奇金淋巴瘤 (non-Hodgkin's lymphoma) 治疗中的结果
Authors Gao Y, Diao L, Li H, Guo Z
Published Date July 2015 Volume 2015:8 Pages 1735—1741
DOI http://dx.doi.org/10.2147/OTT.S86338
Received 9 April 2015, Accepted 11 June 2015, Published 15 July 2015
Objective: microRNA (miRNA)-related single nucleotide polymorphisms (miR-SNPs) in
miRNA-processing machinery genes can affect cancer risk, treatment efficacy,
and patients’ prognosis by mediating the expression of targeted genes. Five
miR-SNPs in miRNA processing machinery genes, including XPO5 (rs11077), RAN (rs14035), TNRC6B (rs9623117), GEMIN3 (rs197412), and GEMIN4 (rs2740348), in 168
non-Hodgkin’s lymphoma (NHL) patients were evaluated for their association with
the cancer risk and outcomes associated with NHL.
Materials and methods: miR-SNPs were genotyped using polymerase chain reaction–ligase detection reaction. The survival curves were calculated using the Kaplan–Meier method, and comparisons between the curves were made using the log-rank test. Multivariate survival analysis was performed using a Cox proportional hazards model.
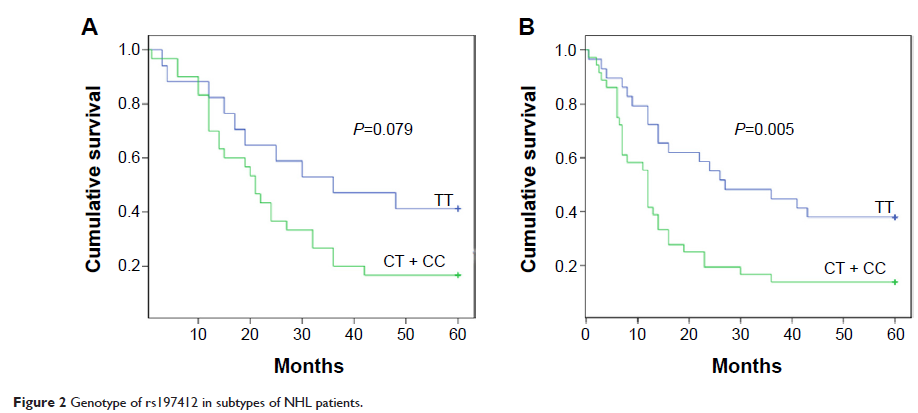
Results: Among the five SNPs, only rs197412 located in the coding region of the GEMIN3 gene was identified; it was independently associated with overall survival in NHL patients, as determined by multivariate analysis (relative risk: 1.649; 95% confidence interval: 1.110–2.449; P =0.013). The prognostic value of this miR-SNP in patient outcomes was also observed in the diffuse large B-cell lymphoma and T-cell lymphoma NHL subtypes.
Conclusion: Our results suggested that the specific genetic variants observed in the miRNA machinery genes may affect NHL survival.
Keywords: miR-SNP, rs197412, GEMIN3 , NHL, survival
Materials and methods: miR-SNPs were genotyped using polymerase chain reaction–ligase detection reaction. The survival curves were calculated using the Kaplan–Meier method, and comparisons between the curves were made using the log-rank test. Multivariate survival analysis was performed using a Cox proportional hazards model.
Results: Among the five SNPs, only rs197412 located in the coding region of the GEMIN3 gene was identified; it was independently associated with overall survival in NHL patients, as determined by multivariate analysis (relative risk: 1.649; 95% confidence interval: 1.110–2.449; P =0.013). The prognostic value of this miR-SNP in patient outcomes was also observed in the diffuse large B-cell lymphoma and T-cell lymphoma NHL subtypes.
Conclusion: Our results suggested that the specific genetic variants observed in the miRNA machinery genes may affect NHL survival.
Keywords: miR-SNP, rs197412, GEMIN3 , NHL, survival