9 1 2 3 6
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 2.6 Breast Cancer (Dove Med Press)
- 3.9 Clin Epidemiol
- 3.3 Cancer Manag Res
- 3.9 Infect Drug Resist
- 3.6 Clin Interv Aging
- 4.8 Drug Des Dev Ther
- 2.8 Int J Chronic Obstr
- 8.0 Int J Nanomed
- 2.3 Int J Women's Health
- 3.2 Neuropsych Dis Treat
- 4.0 OncoTargets Ther
- 2.2 Patient Prefer Adher
- 2.8 Ther Clin Risk Manag
- 2.7 J Pain Res
- 3.3 Diabet Metab Synd Ob
- 4.3 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.9 Pharmgenomics Pers Med
- 3.5 Risk Manag Healthc Policy
- 4.5 J Inflamm Res
- 2.3 Int J Gen Med
- 4.1 J Hepatocell Carcinoma
- 3.2 J Asthma Allergy
- 2.3 Clin Cosmet Investig Dermatol
- 3.3 J Multidiscip Healthc

已发表论文
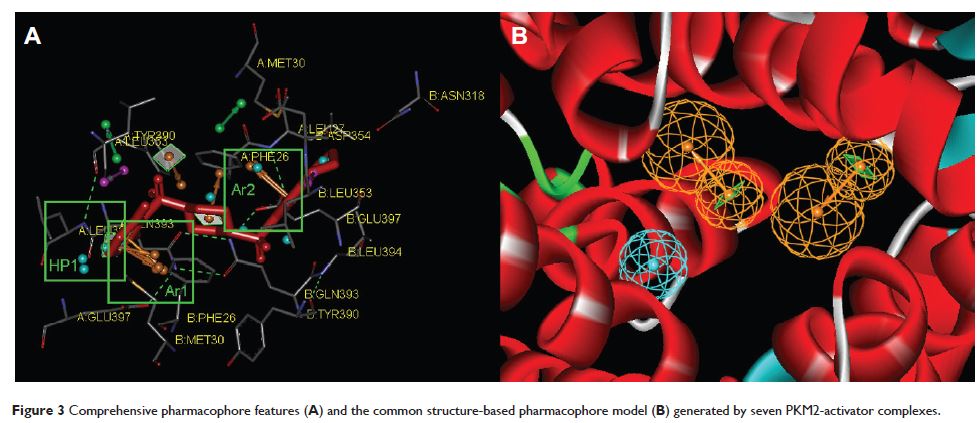
将以结构为基础的药效建模、虚拟筛选以及在硅片 ADMET 分析组合在一起以发现具有抗肿瘤活性的、新的四氢喹啉基丙酮酸激酶同工酶 M2 活化剂
Authors Chen C, Wang T, Wu F, Huang W, He G, Ouyang L, Xiang M, Peng C, Jiang Q
Published Date September 2014 Volume 2014:8 Pages 1195—1210
DOI http://dx.doi.org/10.2147/DDDT.S62921
Received 23 February 2014, Accepted 9 July 2014, Published 2 September 2014
Abstract: Compared with normal differentiated cells, cancer cells upregulate the expression of pyruvate kinase isozyme M2 (PKM2) to support glycolytic intermediates for anabolic processes, including the synthesis of nucleic acids, amino acids, and lipids. In this study, a combination of the structure-based pharmacophore modeling and a hybrid protocol of virtual screening methods comprised of pharmacophore model-based virtual screening, docking-based virtual screening, and in silico ADMET (absorption, distribution, metabolism, excretion and toxicity) analysis were used to retrieve novel PKM2 activators from commercially available chemical databases. Tetrahydroquinoline derivatives were identified as potential scaffolds of PKM2 activators. Thus, the hybrid virtual screening approach was applied to screen the focused tetrahydroquinoline derivatives embedded in the ZINC database. Six hit compounds were selected from the final hits and experimental studies were then performed. Compound 8 displayed a potent inhibitory effect on human lung cancer cells. Following treatment with Compound 8, cell viability, apoptosis, and reactive oxygen species (ROS) production were examined in A549 cells. Finally, we evaluated the effects of Compound 8 on mice xenograft tumor models in vivo. These results may provide important information for further research on novel PKM2 activators as antitumor agents.
Keywords: pharmacophore, molecular docking, pyruvate kinase, virtual screening
Keywords: pharmacophore, molecular docking, pyruvate kinase, virtual screening