9 0 8 0 2
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 2.6 Breast Cancer (Dove Med Press)
- 3.9 Clin Epidemiol
- 3.3 Cancer Manag Res
- 3.9 Infect Drug Resist
- 3.6 Clin Interv Aging
- 4.8 Drug Des Dev Ther
- 2.8 Int J Chronic Obstr
- 8.0 Int J Nanomed
- 2.3 Int J Women's Health
- 3.2 Neuropsych Dis Treat
- 4.0 OncoTargets Ther
- 2.2 Patient Prefer Adher
- 2.8 Ther Clin Risk Manag
- 2.7 J Pain Res
- 3.3 Diabet Metab Synd Ob
- 4.3 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.9 Pharmgenomics Pers Med
- 3.5 Risk Manag Healthc Policy
- 4.5 J Inflamm Res
- 2.3 Int J Gen Med
- 4.1 J Hepatocell Carcinoma
- 3.2 J Asthma Allergy
- 2.3 Clin Cosmet Investig Dermatol
- 3.3 J Multidiscip Healthc

多重耐药肺炎链球菌分离株的比较基因组分析
Authors Pan F, Zhang H, Dong X, Ye W, He P, Zhang SL, Zhu JX, Zhong N
Received 31 July 2017
Accepted for publication 30 January 2018
Published 3 May 2018 Volume 2018:11 Pages 659—670
DOI https://doi.org/10.2147/IDR.S147858
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Colin Mak
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Introduction: Multidrug resistance in Streptococcus pneumoniae has
emerged as a serious problem to public health. A further understanding of the
genetic diversity in antibiotic-resistant S.
pneumoniae isolates is needed.
Methods: We conducted whole-genome resequencing for 25 pneumococcal strains
isolated from children with different antimicrobial resistance profiles.
Comparative analysis focus on detection of single-nucleotide polymorphisms
(SNPs) and insertions and deletions (indels) was conducted. Moreover,
phylogenetic analysis was applied to investigate the genetic relationship among
these strains.
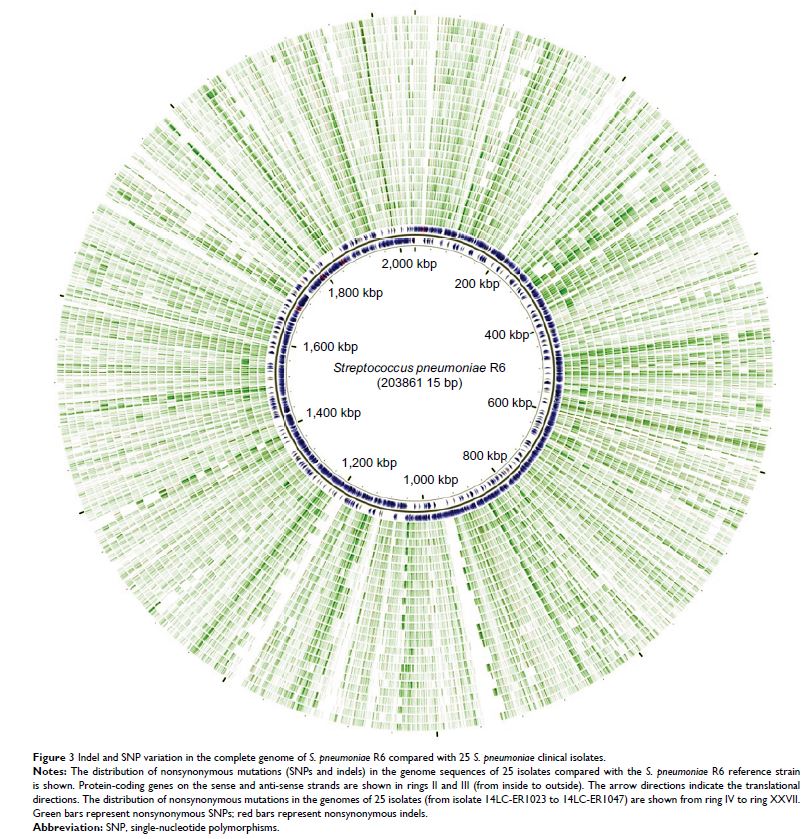
Results: The genome size of the isolates was ~2.1 Mbp, covering >90% of
the total estimated size of the reference genome. The overall G+C% content was
~39.5%, and there were 2,200–2,400 open reading frames. All isolates with
different drug resistance profiles harbored many indels (range 131–171) and
SNPs (range 16,103–28,128). Genetic diversity analysis showed that the
variation of different genes were associated with specific antibiotic
resistance. Known antibiotic resistance genes (pbps , murMN , ciaH , rplD , sulA , and dpr ) were identified, and new genes
(regR , argH , trkH , and PTS-EII ) closely related with
antibiotic resistance were found, although these genes were primarily annotated
with functions in virulence as well as carbohydrate and amino acid transport
and metabolism. Phylogenetic analysis unambiguously indicated that isolates
with different antibiotic resistance profiles harbored similar genetic
backgrounds. One isolate, 14-LC.ER1025, showed a much weaker phylogenetic
relationship with the other isolates, possibly caused by genomic variation.
Conclusion: In this study, although pneumococcal isolates had similar genetic
backgrounds, strains were diverse at the genomic level. These strains exhibited
distinct variations in their indel and SNP compositions associated with drug
resistance.
Keywords: Streptococcus pneumoniae ,
antimicrobial resistance, whole-genome sequencing, insertions/deletions, SNPs,
phylogenetic analysis