9 0 8 1 0
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 2.6 Breast Cancer (Dove Med Press)
- 3.9 Clin Epidemiol
- 3.3 Cancer Manag Res
- 3.9 Infect Drug Resist
- 3.6 Clin Interv Aging
- 4.8 Drug Des Dev Ther
- 2.8 Int J Chronic Obstr
- 8.0 Int J Nanomed
- 2.3 Int J Women's Health
- 3.2 Neuropsych Dis Treat
- 4.0 OncoTargets Ther
- 2.2 Patient Prefer Adher
- 2.8 Ther Clin Risk Manag
- 2.7 J Pain Res
- 3.3 Diabet Metab Synd Ob
- 4.3 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.9 Pharmgenomics Pers Med
- 3.5 Risk Manag Healthc Policy
- 4.5 J Inflamm Res
- 2.3 Int J Gen Med
- 4.1 J Hepatocell Carcinoma
- 3.2 J Asthma Allergy
- 2.3 Clin Cosmet Investig Dermatol
- 3.3 J Multidiscip Healthc

中国东北地区与医院呼吸机相关的肺炎病原菌分布及耐药性分析
Authors Wang Y, Zhang R, Liu W
Received 29 April 2018
Accepted for publication 31 August 2018
Published 13 November 2018 Volume 2018:11 Pages 2249—2255
DOI https://doi.org/10.2147/IDR.S172598
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Colin Mak
Peer reviewer comments 3
Editor who approved publication: Dr Joachim Wink
Purpose: To study
the distribution of pathogenic bacteria in ventilator-associated pneumonia
(VAP), and epidemiological characteristics of extended-spectrum β lactamase
(ESBL)-producing bacteria.
Patients and methods: Lower
respiratory tract secretions from 156 patients with mechanical ventilation were
collected using a protective specimen brush (PSB), with quantitative bacterial
culture carried out and antibiotic sensitivity measured. ESBLs produced by
Gram-negative bacilli were detected using the double disk diffusion method and
monitored by plasmid profiles.
Results: Gram-negative
bacilli accounted for 78.9% of VAP pathogens, with Acinetobacter baumannii (25%), Pseudomonas aeruginosa (19.7%),
and Klebsiella
pneumoniae (14.5%) as the most common strains. There were 12
Gram-positive strains detected (15.8%); mostly methicillin-resistant. Staphylococcus aureus and
methicillin-resistant coagulase-negative Staphylococcus .
There were also four strains of Candida albicans detected (5.26%). Most
Gram-negative bacilli are sensitive to imipenem, but A. baumannii is
serious resistant. ESBLs were detected in nine strains of Gram-negative
bacilli; mainly produced by K. pneumoniae and Escherichia coli ,
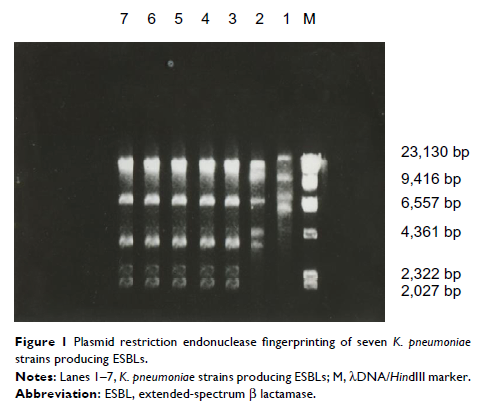
to different degrees of multidrug resistance. Five strains of K. pneumoniae -producing
ESBLs were from the same clonal origin, as confirmed by plasmid restriction
endonuclease analysis.
Conclusion: VAP was mainly
caused by Gram-negative bacteria, with high antibiotic resistance rates.
Plasmids played an important role in the spread of antibiotic resistance among
bacteria.
Keywords: ventilator-associated
pneumonia, pathogenic bacteria, extended-spectrum β lactamases, drug resistance
plasmid