9 0 8 1 0
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 2.6 Breast Cancer (Dove Med Press)
- 3.9 Clin Epidemiol
- 3.3 Cancer Manag Res
- 3.9 Infect Drug Resist
- 3.6 Clin Interv Aging
- 4.8 Drug Des Dev Ther
- 2.8 Int J Chronic Obstr
- 8.0 Int J Nanomed
- 2.3 Int J Women's Health
- 3.2 Neuropsych Dis Treat
- 4.0 OncoTargets Ther
- 2.2 Patient Prefer Adher
- 2.8 Ther Clin Risk Manag
- 2.7 J Pain Res
- 3.3 Diabet Metab Synd Ob
- 4.3 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.9 Pharmgenomics Pers Med
- 3.5 Risk Manag Healthc Policy
- 4.5 J Inflamm Res
- 2.3 Int J Gen Med
- 4.1 J Hepatocell Carcinoma
- 3.2 J Asthma Allergy
- 2.3 Clin Cosmet Investig Dermatol
- 3.3 J Multidiscip Healthc

中国广东省住院患者耐甲氧西林金黄色葡萄球菌菌株的耐药性、毒力基因谱及分子相关性
Authors Liang Y, Tu C, Tan C, El-Sayed Ahmed MAEG, Dai M, Xia Y, Liu Y, Zhong LL, Shen C, Chen G, Tian GB, Liu J, Zheng X
Received 29 October 2018
Accepted for publication 20 December 2018
Published 25 February 2019 Volume 2019:12 Pages 447—459
DOI https://doi.org/10.2147/IDR.S192611
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Cristina Weinberg
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Purpose: The main objective of this study was to
decipher the prevalence, antimicrobial resistance, major virulence genes and
the molecular characteristics of methicillin-resistant Staphylococcus aureus (MRSA)
isolated from different clinical sources in southern China.
Materials and methods: The present study was performed on 187 non-duplicate S. aureus clinical
isolates collected from three tertiary hospitals in Guangdong Province, China,
2010–2016. Antimicrobial susceptibility testing was performed by the disk
diffusion method and by measuring the minimum inhibitory concentration.
Screening for resistance and virulence genes was performed. Clonal relatedness
was determined using various molecular typing methods such as multilocus
sequence typing, spa and staphylococcal chromosomal cassette mec
(SCCmec) typing. Whole genome sequencing was performed for three selected
isolates.
Results: Out
of 187 isolates, 103 (55%) were identified as MRSA. The highest prevalence rate
was found among the skin and soft tissue infection (SSTI) samples (58/103),
followed by sputum samples (25/103), blood stream infection samples (15/103)
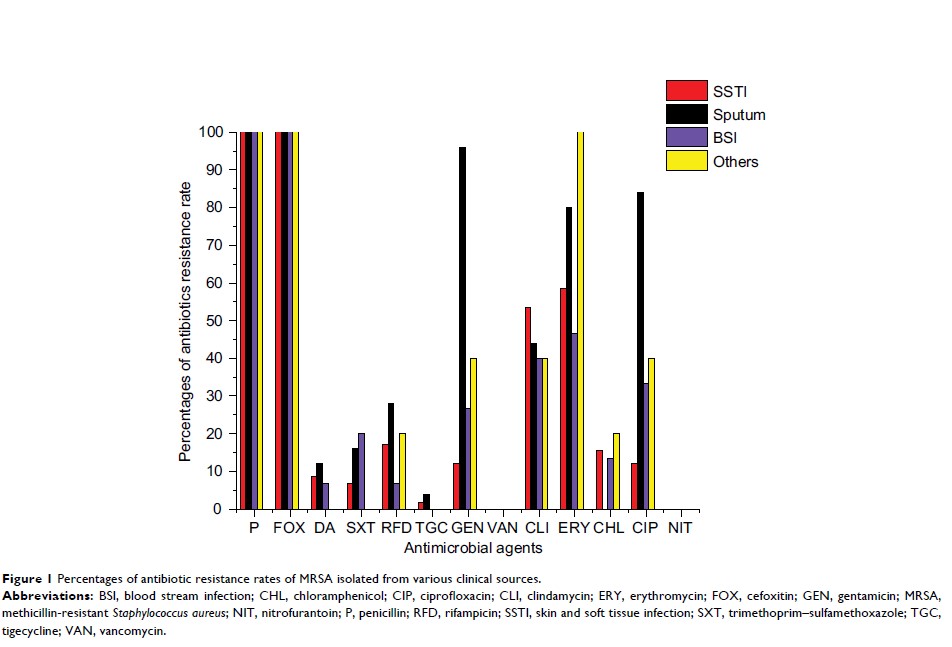
and others (5/103). Antimicrobial susceptibility results revealed high
resistance rates for erythromycin (64.1%), clindamycin (48.5%), gentamicin
(36.9%) and ciprofloxacin (33.98%). All isolates were susceptible to
vancomycin. Resistance genes and mutation detected were as follows: aac(6’)-aph (2”) (24.3%), dfrG (10.7%), rpoB (21.4%), cfr (0%), fexA (1.94%), gyrA (35.92%), gyrB (0.97%), grlA (20.4%), grlB (10.68%), ermA (21.4%), ermB (18.44%), ermC (21.4%)
and lnuA (18.44%).
Profiling of virulence genes revealed the following: sea (11.7%), seb (21.4%), sec (0.97%), sed (0.97%), hla (86.41%), hlb (17.48%), hlg (10.68%), hld (53.4%), Tsst-1 (3.9%)
and pvl (27.2%).
Clonal relatedness showed that ST239-SCCmecA III-t37 clone was the most
prevalent clone.
Conclusion: Our
study elucidated the prevalence, antibiotic resistance, pathogenicity and
molecular characteristics of MRSA isolated from various clinical sources in
Guangdong, China. We found that the infectious rate of MRSA was higher among
SSTI than other sources. The most predominant genotype was ST239-SCCmecA
III-t37 clone, indicating that ST239-t30 clone which was previously predominant
had been replaced by a new clone.
Keywords: MRSA,
antibiotic resistance, resistance genes, virulence factors, molecular typing,
whole genome sequencing