9 0 8 0 2
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 2.6 Breast Cancer (Dove Med Press)
- 3.9 Clin Epidemiol
- 3.3 Cancer Manag Res
- 3.9 Infect Drug Resist
- 3.6 Clin Interv Aging
- 4.8 Drug Des Dev Ther
- 2.8 Int J Chronic Obstr
- 8.0 Int J Nanomed
- 2.3 Int J Women's Health
- 3.2 Neuropsych Dis Treat
- 4.0 OncoTargets Ther
- 2.2 Patient Prefer Adher
- 2.8 Ther Clin Risk Manag
- 2.7 J Pain Res
- 3.3 Diabet Metab Synd Ob
- 4.3 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.9 Pharmgenomics Pers Med
- 3.5 Risk Manag Healthc Policy
- 4.5 J Inflamm Res
- 2.3 Int J Gen Med
- 4.1 J Hepatocell Carcinoma
- 3.2 J Asthma Allergy
- 2.3 Clin Cosmet Investig Dermatol
- 3.3 J Multidiscip Healthc

已发表论文
针对结直肠癌的潜在的诊断标志物和治疗靶点的筛选
Authors Tian XQ, Sun DF, Zhao SL, Xiong H, Fang JY
Published Date July 2015 Volume 2015:8 Pages 1691—1699
DOI http://dx.doi.org/10.2147/OTT.S81621
Received 26 January 2015, Accepted 16 March 2015, Published 8 July 2015
Objective: To identify genes with aberrant promoter methylation for developing
novel diagnostic markers and therapeutic targets against primary colorectal
cancer (CRC).
Methods: Two paired CRC and adjacent normal tissues were collected from two CRC patients. A Resi: MBD2b protein-sepharose-4B column was used to enrich the methylated DNA fragments. Difference in the average methylation level of each DNA methylation region between the tumor and control samples was determined by log2 fold change (FC) in each patient to screen the differentially methylated DNA regions. Genes with log2FC value ≥4 or ≤-4 were identified to be hypermethylated and hypomethylated, respectively. Then, the underlying functions of methylated genes were speculated by Gene Ontology database and pathway enrichment analyses. Furthermore, a protein–protein interaction network was built using Search Tool for the Retrieval of Interacting Genes/Proteins database, and the transcription factor binding sites were screened via the Encyclopedia of DNA Elements (ENCODE) database.
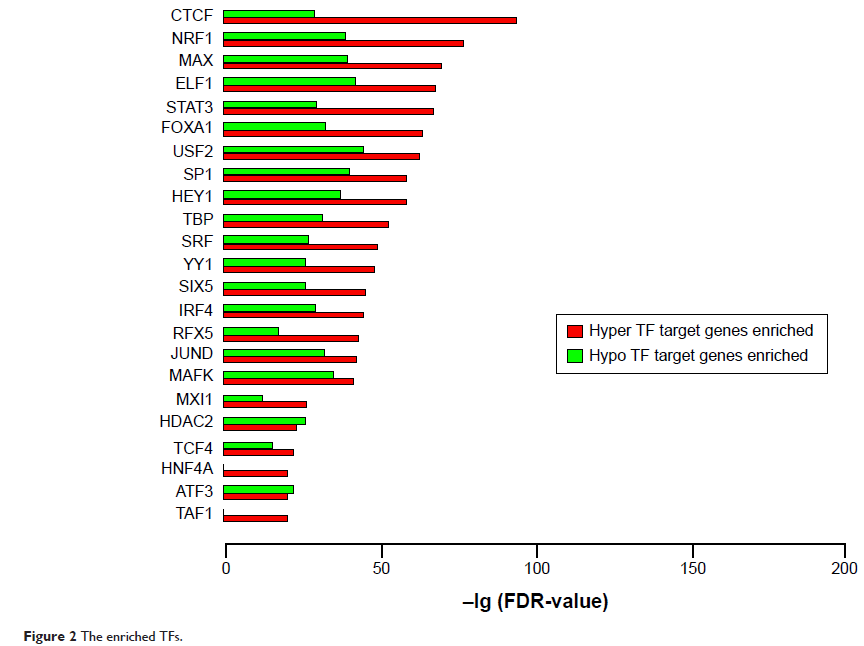
Results: Totally, 2,284 and 1,142 genes were predicted to have aberrant promoter hypermethylation or hypomethylation, respectively. MAP3K5 , MAP3K8 , MAPK14 , and MAPK9 with promoter hypermethylation functioned via MAPK signaling pathway, focal adhesion, or Wnt signaling pathway, whereas MAP2K1 , MAPK3 , MAPK11 , and MAPK7 with promoter hypomethylation functioned via TGF-beta signaling pathway, neurotrophin signaling pathway, and chemokine signaling pathway. CREBBP, PIK3R1, MAPK14, APP, ESR1, MAPK3, and HRAS were the seven hubs in the constructed protein–protein interaction network. RPL22 , RPL36 , RPLP2 , RPS7 , and RPS9 were commonly regulated by transcription factors, and YY1 and IRF4 were hypermethylated.
Conclusion: MAPK14 , MAPK3 , HRAS , YY1 , and IRF4 may be considered as potential biomarkers for early diagnosis and therapy of CRC.
Keywords: primary colorectal cancer, aberrant DNA methylation, microarray analysis, pathway enrichment analysis, transcription factor
Methods: Two paired CRC and adjacent normal tissues were collected from two CRC patients. A Resi: MBD2b protein-sepharose-4B column was used to enrich the methylated DNA fragments. Difference in the average methylation level of each DNA methylation region between the tumor and control samples was determined by log2 fold change (FC) in each patient to screen the differentially methylated DNA regions. Genes with log2FC value ≥4 or ≤-4 were identified to be hypermethylated and hypomethylated, respectively. Then, the underlying functions of methylated genes were speculated by Gene Ontology database and pathway enrichment analyses. Furthermore, a protein–protein interaction network was built using Search Tool for the Retrieval of Interacting Genes/Proteins database, and the transcription factor binding sites were screened via the Encyclopedia of DNA Elements (ENCODE) database.
Results: Totally, 2,284 and 1,142 genes were predicted to have aberrant promoter hypermethylation or hypomethylation, respectively. MAP3K5 , MAP3K8 , MAPK14 , and MAPK9 with promoter hypermethylation functioned via MAPK signaling pathway, focal adhesion, or Wnt signaling pathway, whereas MAP2K1 , MAPK3 , MAPK11 , and MAPK7 with promoter hypomethylation functioned via TGF-beta signaling pathway, neurotrophin signaling pathway, and chemokine signaling pathway. CREBBP, PIK3R1, MAPK14, APP, ESR1, MAPK3, and HRAS were the seven hubs in the constructed protein–protein interaction network. RPL22 , RPL36 , RPLP2 , RPS7 , and RPS9 were commonly regulated by transcription factors, and YY1 and IRF4 were hypermethylated.
Conclusion: MAPK14 , MAPK3 , HRAS , YY1 , and IRF4 may be considered as potential biomarkers for early diagnosis and therapy of CRC.
Keywords: primary colorectal cancer, aberrant DNA methylation, microarray analysis, pathway enrichment analysis, transcription factor