111314
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

小鼠嗅觉功能障碍期间代谢网络的共生微生物群调节
Authors Wang H, Liu L, Rao X, Chai T, Zeng B, Zhang X, Yu Y, Zhou C, Pu J, Zhou W, Li W, Zhang H, Wei H, Xie P
Received 30 October 2019
Accepted for publication 1 March 2020
Published 19 March 2020 Volume 2020:16 Pages 761—769
DOI https://doi.org/10.2147/NDT.S236541
Checked for plagiarism Yes
Review by Single-blind
Peer reviewer comments 3
Editor who approved publication: Professor Jun Chen
Introduction: Recently, an increasing number of studies have focused on commensal microbiota. These microorganisms have been suggested to impact human health and disease. However, only a small amount of data exists to support the assessment of the influences that commensal microbiota exert on olfactory function.
Methods: We used a buried food pellet test (BFPT) to investigate and compare olfactory functions in adult, male, germ-free (GF) and specific-pathogen-free (SPF) mice, then examined and compared the metabolomic profiles for olfactory bulbs (OBs) isolated from GF and SPF mice to uncover the mechanisms associated with olfactory dysfunction.
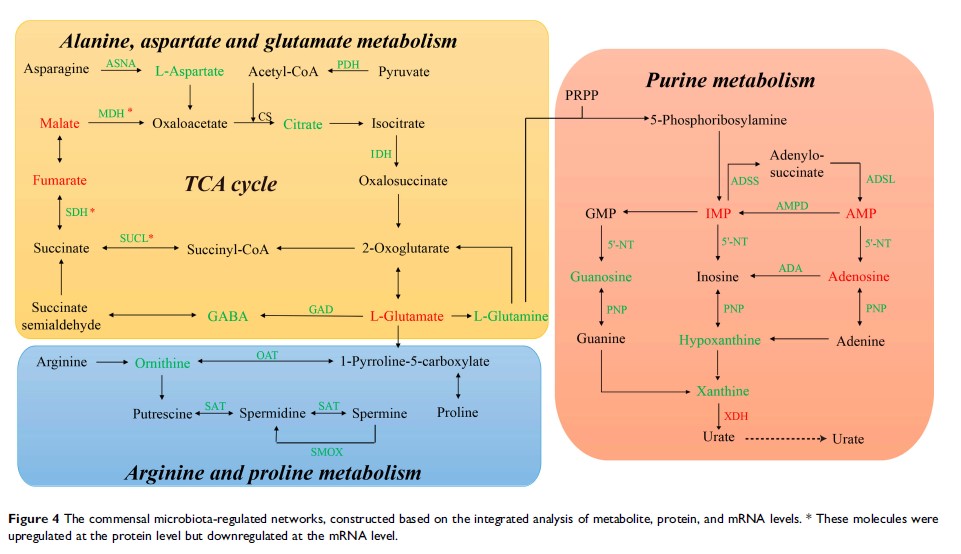
Results: We found that the absence of commensal microbiota was able to influence olfactory function and the metabolic signatures of OBs, with 38 metabolites presenting significant differences between the two groups. These metabolites were primarily associated with disturbances in glycolysis, the tricarboxylic acid (TCA) cycle, amino acid metabolism, and purine catabolism. Finally, the commensal microbiota regulation of metabolic networks during olfactory dysfunction was identified, based on an integrated analysis of metabolite, protein, and mRNA levels.
Conclusion: This study demonstrated that the absence of commensal microbiota may impair olfactory function and disrupt metabolic networks. These findings provide a new entry-point for understanding olfactory-associated disorders and their potential underlying mechanisms.
Keywords: gut microbiota, germ-free, olfactory bulb, metabolomic, gas chromatography-mass spectrometry