111314
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

新生儿重症监护病房暴发产超广谱 β-内酰胺酶肺炎克雷伯菌 ST471 的基因组流行病学
Authors Wang Y, Luo C, Du P, Hu J, Zhao X, Mo D, Du X, Xu X, Li M, Lu H, Zhou Z, Cui Z, Zhou H
Received 26 October 2019
Accepted for publication 11 January 2020
Published 15 April 2020 Volume 2020:13 Pages 1081—1090
DOI https://doi.org/10.2147/IDR.S236212
Checked for plagiarism Yes
Review by Single-blind
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Purpose: Klebsiella pneumoniae producing extended-spectrum β-lactamases (ESBLs) causes nosocomial infections worldwide. The present study aimed to determine the molecular subtyping characteristics and antibiotic resistance mechanisms of ESBL-producing K. pneumoniae strains collected during an outbreak. Moreover, we attempted to reveal the fine transmission route of the strains within this outbreak using whole-genome sequencing (WGS).
Methods: Collecting cases and strain information were carried out. Outbreak-related strains were identified using pulsed-field gel electrophoresis (PFGE). The antibiotic susceptibility, drug-resistant genes, and molecular subtype characteristics of ESBL-producing K. pneumoniae were analyzed. The fine transmission route of the strains within this outbreak was revealed using WGS and minimum core genome (MCG) sequence typing.
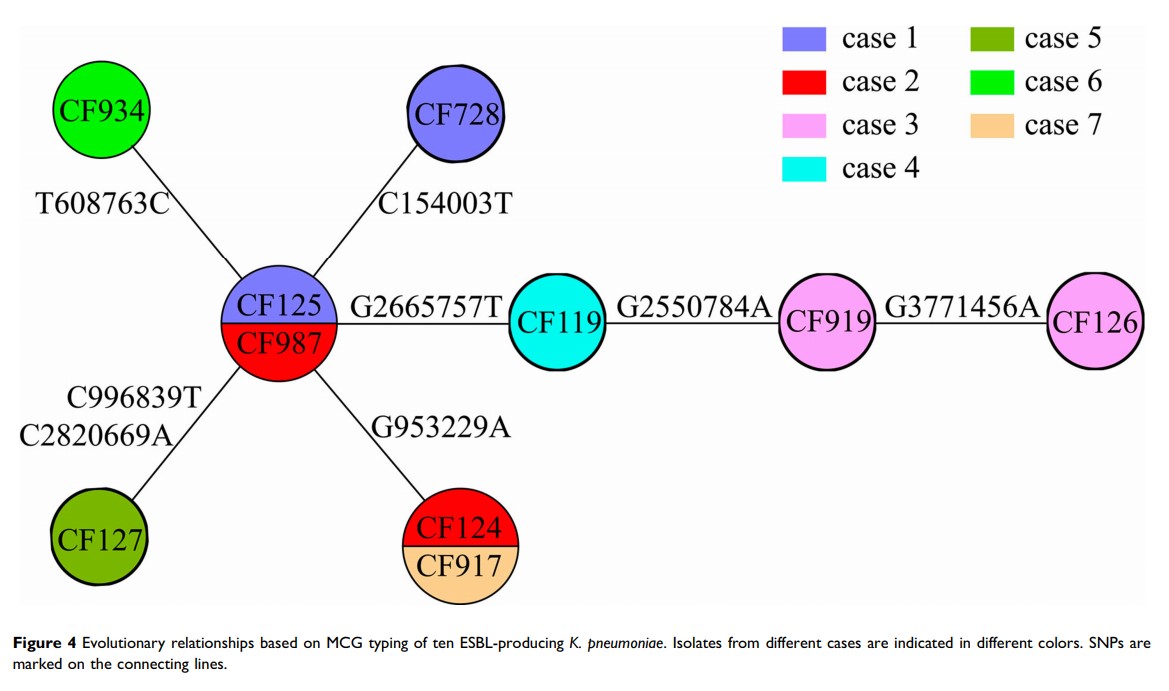
Results: In mid-January, 2015, five cases of neonatal pneumonia caused by ESBL-producing K. pneumoniae were observed in the neonatal intensive care unit (NICU) of the Affiliated Hospital of Chifeng University, China. Eight ESBL-producing K. pneumoniae were isolated from these five cases, and two additional strains from another two cases were identified using PFGE. All ten isolates harbored bla CTX-M-15, bla TEM-1, bla SHV-108, and bla OXA-1 genes, and belonged to the sequence type 471 (ST471) clone. A putative transmission map was constructed via comprehensive consideration of genomic and epidemiological information. WGS identified the initial case and the “superspreader”. The genomic epidemiological investigation revealed that the outbreak was caused by the introduction of the bacteria one month before the first case appeared.
Conclusion: As far as we know, this is the first report to describe the characteristics of an ST471 ESBL-producing K. pneumoniae outbreak. The data showed that epidemiological inferences could be greatly improved by interpretation in the context of WGS and that K. pneumoniae strains isolated from the same outbreak contain sufficient genomic differences to refine epidemiological linkages on the basis of genetic lineage. These findings suggested that integration of genomic and epidemiological data can help us to have a clearer understanding of when and how outbreaks occur, so as to better control nosocomial transmission.
Keywords: whole-genome sequencing, minimum core genome, superspreader, transmission map