111314
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

Patients with NSCLCs Harboring Internal Inversions or Deletion Rearrangements of the ALK Gene Have Durable Responses to ALK Kinase Inhibitors
Authors Schrock AB, Madison R, Rosenzweig M, Allen JM, Erlich RL, Wang SY, Chidiac T, Reddy VS, Riess JW, Yassa AE, Shakir A, Miller VA, Alexander BM, Venstrom J, McGregor K, Ali SM
Received 22 November 2019
Accepted for publication 24 March 2020
Published 17 April 2020 Volume 2020:11 Pages 33—39
DOI https://doi.org/10.2147/LCTT.S239675
Checked for plagiarism Yes
Review by Single-blind
Peer reviewer comments 2
Editor who approved publication: Dr Sai-Hong Ignatius Ou
Background: ALK fusions are targetable drivers in non-small-cell lung cancer (NSCLC). However, patients with NSCLC harboring ALK rearrangements without a fusion partner identified in DNA have also been shown to respond to ALK inhibitors. We aimed to characterize complex ALK variants that may predict sensitivity to multiple approved ALK inhibitors.
Methods: Comprehensive genomic profiling (CGP) of DNA isolated from formalin‐fixed paraffin‐embedded (FFPE) tumor tissue or blood-based circulating tumor DNA was performed for 39,159 NSCLC patients during routine clinical care. For a subset of cases, RNA sequencing was performed, and prior ALK test results and clinical treatment information were collected from treating physicians.
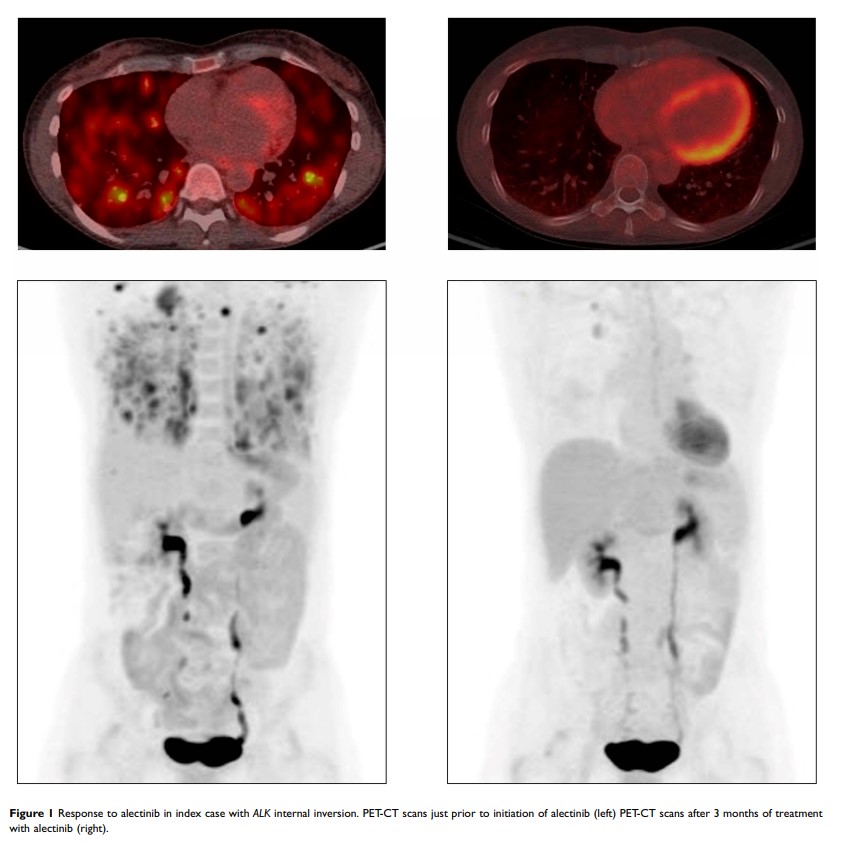
Results: We queried the Foundation Medicine NSCLC database and identified ALK internal inversions, as well as internal deletions, as the sole ALK rearrangements in 6 (0.02%) and 3 (0.01%) of cases, respectively. In cases with ALK internal inversions, RNA testing identified an EML4-ALK fusion in 2/2 cases evaluated, and 3/3 patients treated with ALK inhibitors had durable responses. A single patient with an ALK internal deletion and clinical data available responded to multiple ALK inhibitors. RNA data available for a subset of non-NSCLC cases suggest that ALK internal deletions removing a portion of the N-terminus are drivers themselves and do not result in ALK fusions. Fluorescence in situ hybridization (FISH) results were inconsistent for both classes of DNA events.
Conclusion: Rare internal inversions of ALK appear to be indicative of ALK fusions, which can be detected in RNA, and response to ALK inhibitors in patients with NSCLC. In contrast, ALK internal deletions are not associated with ALK fusions in RNA but likely represent targetable drivers themselves. These data suggest that CGP of DNA should be supplemented with immunohistochemistry or RNA-based testing to further resolve these events and match patients to effective therapies.
Keywords: ALK rearrangement, inversion, deletion, genomic profiling, targeted therapy