110932
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

lncRNA HOTTIP rs3807598 C>G 与先天性巨结肠症(希尔施普龙病)呈负相关
Authors Zheng Y, Zhuo Z, Xie X, Lu L, He Q, Zhong W
Received 13 February 2020
Accepted for publication 22 April 2020
Published 6 May 2020 Volume 2020:13 Pages 151—156
DOI https://doi.org/10.2147/PGPM.S249649
Checked for plagiarism Yes
Review by Single-blind
Peer reviewer comments 2
Editor who approved publication: Dr Martin H. Bluth
Background: Hirschsprung disease (HSCR) is a congenital disease that arises from defective intestinal neural system. LncRNA HOTTIP is a critical gene in various diseases, including HSCR. No epidemiological studies have explored the correlation between lncRNA HOTTIP single nucleotide polymorphisms (SNPs) and HSCR risk. We here lead as a pioneer to explore whether SNPs in lncRNA HOTTIP impact the risk of HSCR and HSCR subtypes in an unrelated Chinese population.
Methods: We used the TaqMan method to genotype rs3807598 C>G of the lncRNA HOTTIP gene using 1470 HSCR cases and 1473 healthy controls. Of them, 1441 cases and 1434 controls were successfully genotyped. We adopted odds ratios (ORs) and 95% confidence intervals (CIs) to quantify the relationship.
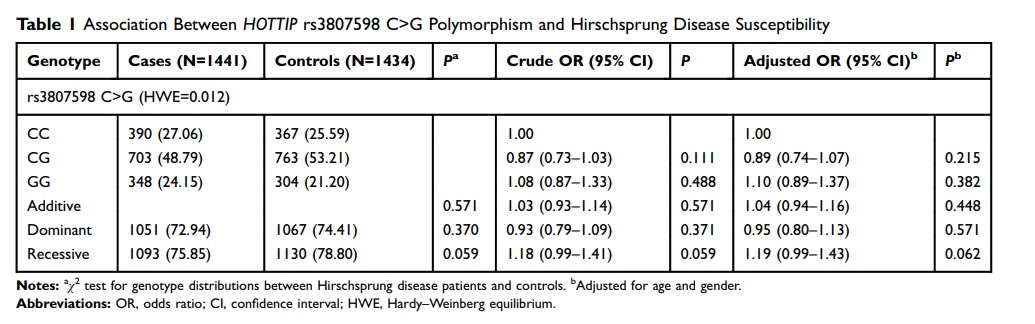
Results: We got an unexpected outcome that lncRNA HOTTIP SNP rs3807598 C>G could not modify the risk of HSCR (CG vs. CC: adjusted OR=0.89, 95% CI=0.74– 1.07; GG vs. CC: adjusted OR=1.10, 95% CI=0.89– 1.37; GG/CG vs CC: adjusted OR=0.95, 95% CI=0.80– 1.13; and GG vs. CC/CG: adjusted OR=1.19, 95% CI=0.99– 1.43). What’s more, risk effect of lncRNA HOTTIP rs3807598 C>G is still not obvious in stratification analysis by HSCR subtype.
Conclusion: Our studies did not provide statistical evidence of a correlation between lncRNA HOTTIP SNP rs3807598 C>G and susceptibility of HSCR in the Chinese population that is being studied. Further validation study with a larger sample size covering multi-ethnic groups is warranted.
Keywords: Hirschsprung disease, HOTTIP , polymorphism, susceptibility, Chinese