110932
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

中国湘雅医院尿结石患者连续五年的尿病原菌培养及药物敏感性分析
Authors Bai Y, Liu Q, Gu J, Zhang X, Hu S
Received 4 December 2019
Accepted for publication 23 April 2020
Published 11 May 2020 Volume 2020:13 Pages 1357—1363
DOI https://doi.org/10.2147/IDR.S241036
Checked for plagiarism Yes
Review by Single-blind
Peer reviewer comments 2
Editor who approved publication: Dr Eric Nulens
Objective: To analyze pathogen distribution and drug sensitivity in patients with urinary calculi and thereby gain insight into the most appropriate antibacterial drugs for perioperative therapy.
Materials and Methods: From January 2014 to December 2018, the results of mid-stream urine pathogen culture and drug sensitivity tests were evaluated retrospectively for 353 patients with urinary calculi. SPSS software version 23.0 was used to analyze the data.
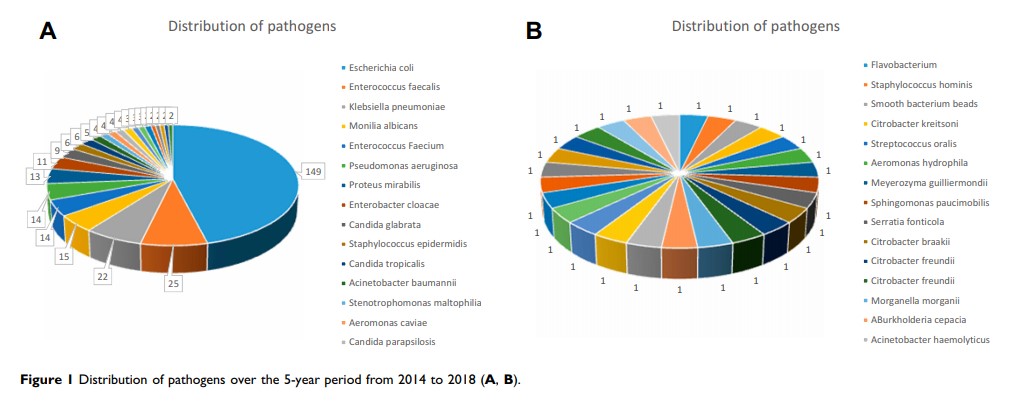
Results: A total of 353 strains of pathogens were isolated from urine culture. Among these, 278 (79%) strains belonged to the top 10 most frequently isolated pathogens, comprising 209 (75.2%) Gram-negative bacilli and 69 (24.8%) Gram-positive cocci. Escherichia coli was the most frequently isolated pathogen overall and the most frequently isolated Gram-negative bacillus, and Enterococcus faecalis was the most frequently isolated Gram-positive coccus. Drug sensitivity levels were effectively unchanged for less commonly used drugs, whereas drug resistance rates remained high for commonly used drugs such as ampicillin trihydrate, ampicillin/sulbactam, cefazolin, ceftriaxone, aztreonam, levofloxacin and ciprofloxacin.
Conclusion: E. coli and E. faecalis remain the most common Gram-negative bacillus and Gram-positive coccus uropathogens, respectively, in patients with urinary calculi. Mid-stream urine pathogen culture and drug sensitivity tests should be used to select appropriate antibacterial drugs before treatment, particularly for perioperative patients with urinary calculi.
Keywords: urine pathogen culture, drug sensitivity, urinary calculi, Escherichia coli, Enterococcus faecalis