110932
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

基于 GEO 和 TCGA 数据库确定炎性乳腺癌潜在关键基因和途径
Authors Lv Q, Liu Y, Huang H, Zhu M, Wu J, Meng D
Received 24 March 2020
Accepted for publication 18 May 2020
Published 12 June 2020 Volume 2020:13 Pages 5541—5550
DOI https://doi.org/10.2147/OTT.S255300
Checked for plagiarism Yes
Review by Single-blind
Peer reviewer comments 3
Editor who approved publication: Dr Federico Perche
Introduction: Inflammatory breast cancer (IBC) is a rare type of breast cancer with poor prognosis, and the pathogenesis of this life-threatening disease is yet to be fully elucidated. This study aims to identify key genes of IBC, which could be potential diagnostic or therapeutic targets.
Methods: Four datasets GSE5847, GSE22597, GSE23720, and GSE45581 were downloaded from the Gene Expression Omnibus (GEO) and differential expression analysis was performed. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were conducted to understand the potential bio-functions of the differentially expressed genes (DEGs). Protein–protein interaction (PPI) network was constructed for functional modules analysis and hub genes identification, and TCGA survival analysis and qRT-PCR of clinical samples were used to further explore and validate the effect of hub genes on IBC.
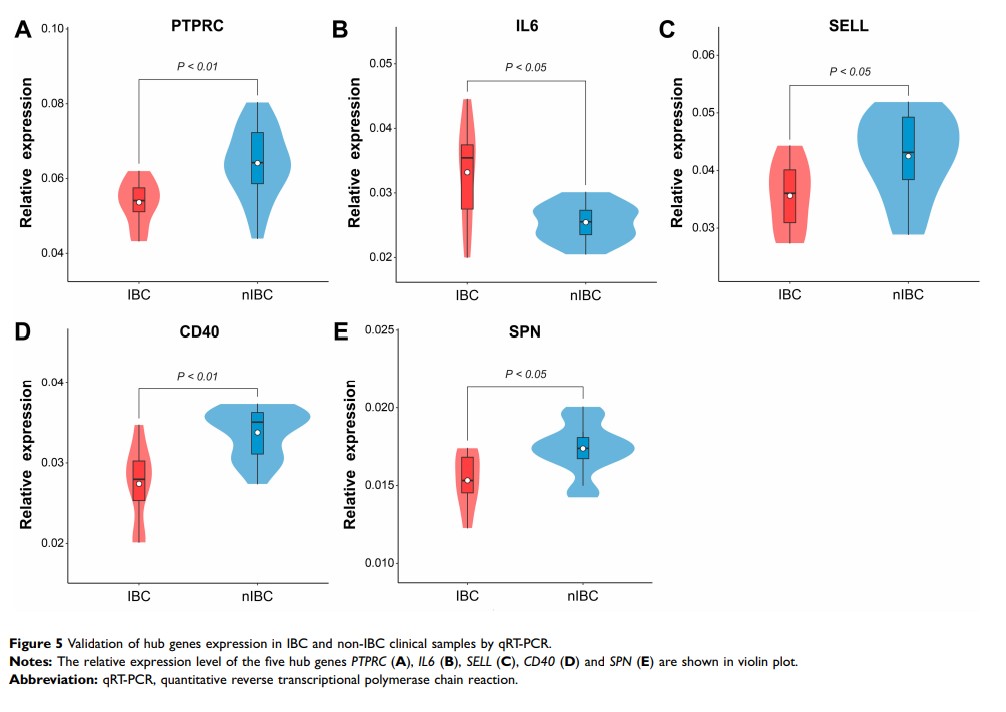
Results: A total of 114 DEGs were identified from the GEO datasets. GO and KEGG analyses showed that the DEGs were mainly enriched in oncogenesis and cell adhesion. From the PPI network, we screened out five hub genes, including PTPRC, IL6, SELL, CD40 , and SPN . Survival analysis and expression validation verified the robustness of the hub genes.
Discussion: The present study provides new insight into the understanding of IBC pathogenesis and the identified hub genes may serve as potential targets for diagnosis and treatment.
Keywords: inflammatory breast cancer, bioinformatic analysis, hub genes, microarray