110932
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

通过整合加权基因共表达网络分析和免疫组织化学验证,PTGDS 表达的降低可预测子宫内膜癌患者的低生存率
Authors Zou R, Zheng M, Tan M, Xu H, Luan N, Zhu L
Received 27 March 2020
Accepted for publication 4 June 2020
Published 26 June 2020 Volume 2020:12 Pages 5057—5075
DOI https://doi.org/10.2147/CMAR.S255753
Checked for plagiarism Yes
Review by Single-blind
Peer reviewer comments 2
Editor who approved publication: Dr Yong Teng
Purpose: To identify key pathogenic genes and reveal the potential molecular mechanisms of endometrial cancer (EC) using bioinformatics analysis and immunohistochemistry validation.
Materials and Methods: Through weighted gene co-expression network analysis (WGCNA), a co-expression network was constructed based on the top 25% variant genes in the GSE50830 dataset downloaded from gene expression omnibus (GEO). GO and KEGG pathway enrichment analyses were performed using the DAVID online tool. Candidate genes were selected using the cytoHubba plug-in of Cytoscape, mRNA expression levels and prognostic values in EC were analyzed by Oncomine, GEPIA, and Kaplan–Meier Plotter database to determine hub genes. One hub gene was validated by immunohistochemical (IHC) staining of 116 paraffin-embedded endometrial tissues and TCGA-UCEC cohort. Genes co-expressed with this hub gene were identified by LinkedOmics. Finally, its correlation with immune infiltration was evaluated by TIMER.
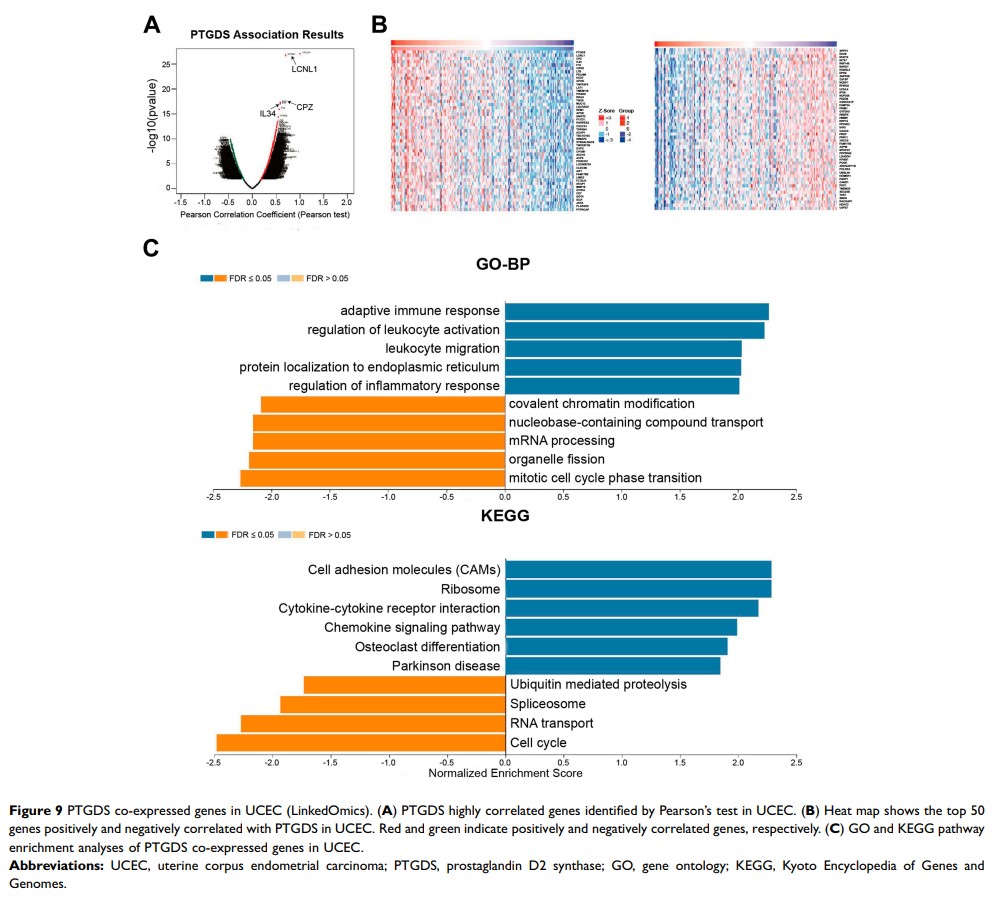
Results: Three co-expression modules and five candidate genes in each module were obtained by WGCNA; four hub genes were identified (LGR5, SST, ZNF558, and PTGDS). The mRNA levels of LGR5 and SST were significantly upregulated in EC, whereas those of ZNF558 and PTGDS were significantly downregulated; the expression of all four genes was associated with EC prognosis. Further validation demonstrated that PTGDS was significantly downregulated in the EC group compared with the atypical hyperplasia and normal endometrial groups, and its low expression was an independent risk factor for worse prognosis of EC. Biological function analysis indicated that PTGDS might be involved in the adaptive immune response, leukocyte migration, as well as in the regulation of cell adhesion molecules and chemokine signaling. Additionally, PTGDS expression was positively correlated with immune infiltration status of B cells, CD4+ T cells and macrophages.
Conclusion: LGR5, SST, ZNF558, and PTGDS may participate in the development, progression, and prognosis of EC, in which PTGDS may be a novel biomarker and therapeutic target for EC.
Keywords: endometrial cancer, bioinformatics analysis, WGCNA, immunohistochemistry, PTGDS