110932
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

使用随机森林模型鉴定的口腔鳞状细胞癌中黏膜微生物组的特征
Authors Zhou J, Wang L, Yuan R, Yu X, Chen Z, Yang F, Sun G, Dong Q
Received 24 February 2020
Accepted for publication 12 June 2020
Published 3 July 2020 Volume 2020:12 Pages 5353—5363
DOI https://doi.org/10.2147/CMAR.S251021
Checked for plagiarism Yes
Review by Single-blind
Peer reviewer comments 3
Editor who approved publication: Dr Antonella D'Anneo
Objective: The aim of this study was to explore the signatures of oral microbiome associated with OSCC using a random forest (RF) model.
Patients and Methods: A total of 24 patients with OSCC were enrolled in the study. The oral microbiome was assessed in cancerous lesions and matched paracancerous tissues from each patient using 16S rRNA gene sequencing. Signatures of mucosal microbiome in OSCC were identified using a RF model.
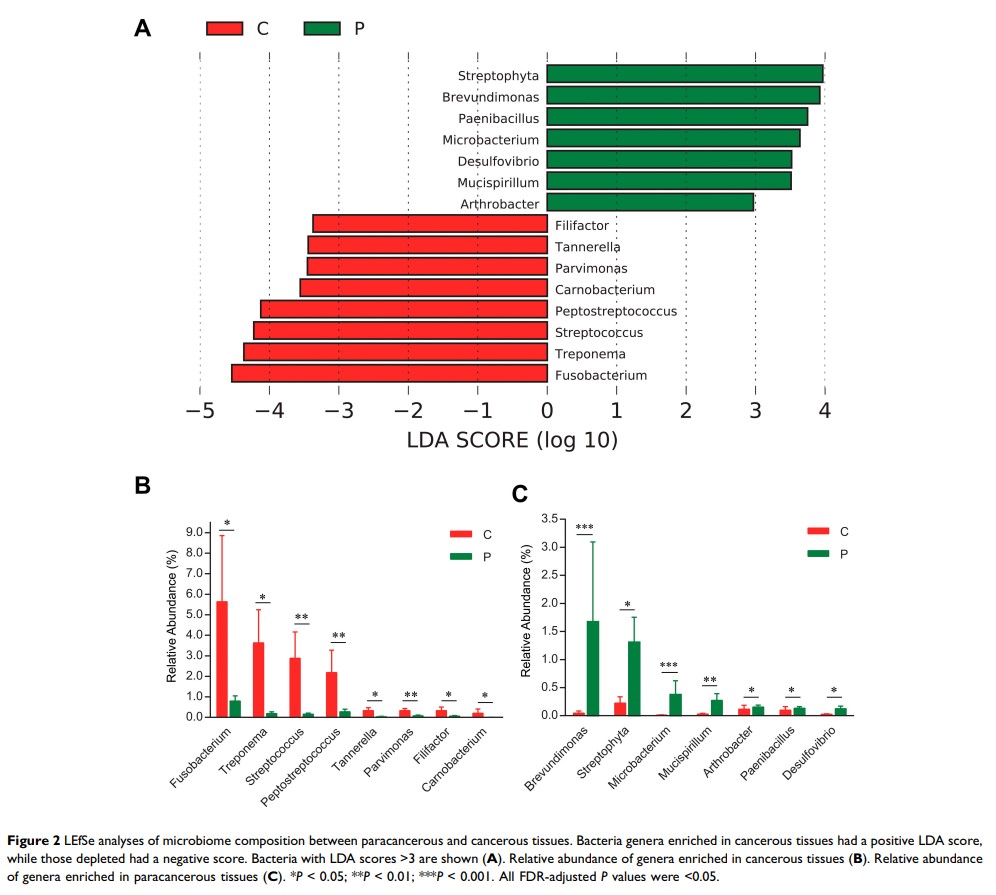
Results: Significant differences were found between OSCC lesions and matched paracancerous tissues with respect to the microbial profile and composition. Linear discriminant analysis effect size analyses (LEfSe) identified 15 bacteria genera associated with cancerous lesions. Fusobacterium, Treponema, Streptococcus, Peptostreptococcus, Carnobacterium, Tannerella, Parvimonas and Filifactor were enriched. A classifier based on RF model identified a microbial signature comprising 12 bacteria, which was capable of distinguishing cancerous lesions and paracancerous tissues (AUC = 0.82). The network of the oral microbiome in cancerous lesions appeared to be simplified and fragmented. Functional analyses of oral microbiome showed altered functions in amino acid metabolism and increased capacity of glucose utilization in OSCC.
Conclusion: The identified microbial signatures may potentially be used as a biomarker for predicting OSCC or for clinical assessment of oral cancer risk.
Keywords: oral squamous cell carcinoma, microbiome, random forest machine learning, predicted functions