108605
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

基于 iTRAQ 的蛋白质组学揭示了产 ESBL 的肺炎克雷伯菌的潜在抗毒力靶标
Authors Wang Y, Cong S, Zhang Q, Li R, Wang K
Received 2 May 2020
Accepted for publication 28 July 2020
Published 19 August 2020 Volume 2020:13 Pages 2891—2899
DOI https://doi.org/10.2147/IDR.S259894
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Purpose: Treatment of infections with Klebsiella pneumoniae strains producing extended-spectrum beta-lactamases (ESBLs) is challenging due to the coexistence of multiple resistance mechanisms and the hypervirulent variant. Therefore, new targets or more effective treatment options aimed at ESBL-producing Klebsiella pneumoniae are urgently needed.
Materials and Methods: Here, we collected ESBL-producing and non-ESBL Klebsiella pneumoniae isolates and studied their differences from a proteomic point of view.
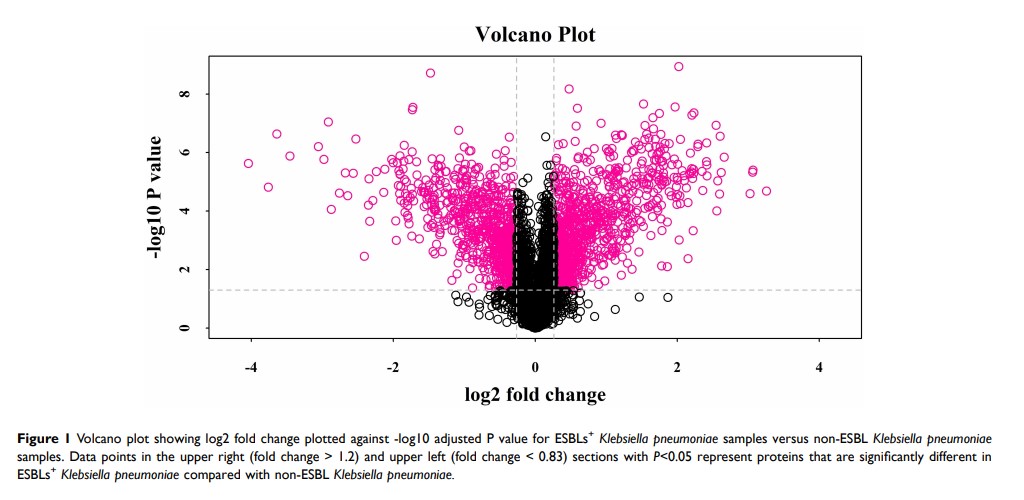
Results: We revealed treA, wza, gnd, rmlA, rmlC, rmlD, galE, aceE, and sucD as important virulence-related proteins in ESBL-producing Klebsiella pneumoniae , distinct from those in non-ESBL strains.
Conclusion: Our findings provide plausible anti-virulence targets and suggest new therapeutic avenues against ESBL-producing Klebsiella pneumoniae .
Keywords: Klebsiella pneumoniae , extended-spectrum beta-lactamases, proteomics, anti-virulence