108605
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

整合基因组分析揭示了对酪氨酸激酶抑制剂耐药或不耐受的慢性髓细胞白血病患者与癌症相关的基因突变
Authors Wu W, Xu N, Zhou X, Liu L, Tan Y, Luo J, Huang J, Qin J, Wang J, Li Z, Yin C, Zhou L, Liu X
Received 10 April 2020
Accepted for publication 30 July 2020
Published 25 August 2020 Volume 2020:13 Pages 8581—8591
DOI https://doi.org/10.2147/OTT.S257661
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr XuYu Yang
Introduction: While the acquisition of mutations in the ABL1 kinase domain (KD) has been identified as a common mechanism behind tyrosine kinase inhibitor (TKI) resistance, recent genetic studies have revealed that patients with TKI resistance or intolerance frequently harbor one or more genetic alterations implicated in myeloid malignancies. This suggests that additional mutations other than ABL1 KD mutations might contribute to disease progression.
Methods: We performed targeted-capture sequencing of 127 known and putative cancer-related genes of 63 patients with CML using next-generation sequencing (NGS), including 42 patients with TKI resistance and 21 with TKI intolerance.
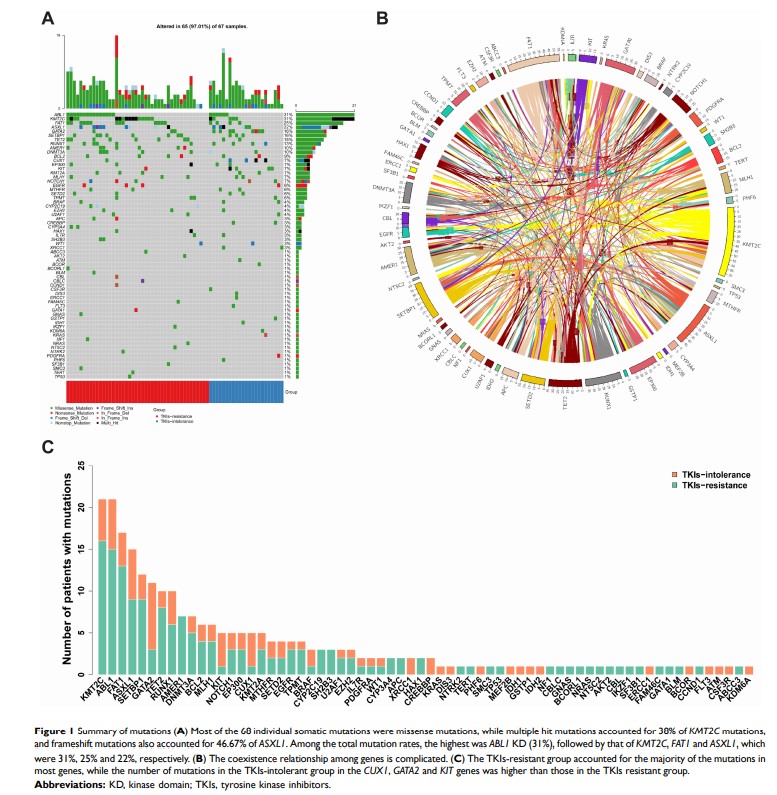
Results: The differences in the number of mutations between groups had no statistical significance. This could be explained in part by not all of the patients having achieved major molecular remission in the early period as expected. Overall, 66 mutations were identified in 96.8% of the patients, most frequently in the KTM2C (31.82%), ABL1 (31.82%), FAT1 (25.76%), and ASXL1 (22.73%) genes. CUX1 , KIT , and GATA2 were associated with TKI intolerance, and two of them (CUX1 , GATA2 ) are transcription factors in which mutations were identified in 82.61% of patients with TKI intolerance. ASXL1 mutations were found more frequently in patients with ABL1 KD mutations (38.1% vs 15.21%, P=0.041). Although the number of mutations was low, pairwise interaction between mutated genes showed that ABL1 KD mutations cooccurred with SH2B3 mutations (P< 0.05). In Kaplan–Meier analyses, only TET2 mutations were associated with shorter progression-free survival (P=0.026).
Conclusion: Our data suggested that the CUX1 , KIT , and GATA2 genes may play important roles in TKI intolerance. ASXL1 and TET2 mutations may be associated with poor patient prognosis. NGS helps improving the clinical risk stratification, which enables the identification of patients with TKI resistance or intolerance in the era of TKI therapy.
Keywords: chronic myeloid leukemia, mutations, tyrosine kinase inhibitor, intolerance, resistance