108605
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

确定 G2 和 S 相表达-1 作为前列腺癌患者的潜在生物标志物
Authors Xiong J, Zhang J, Li H
Received 19 July 2020
Accepted for publication 28 August 2020
Published 28 September 2020 Volume 2020:12 Pages 9259—9269
DOI https://doi.org/10.2147/CMAR.S272795
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Seema Singh
Background: This study aimed to predict and explore the possible clinical value and mechanism of genetic markers in prostate cancer (PCa) using a bioinformatics analysis method.
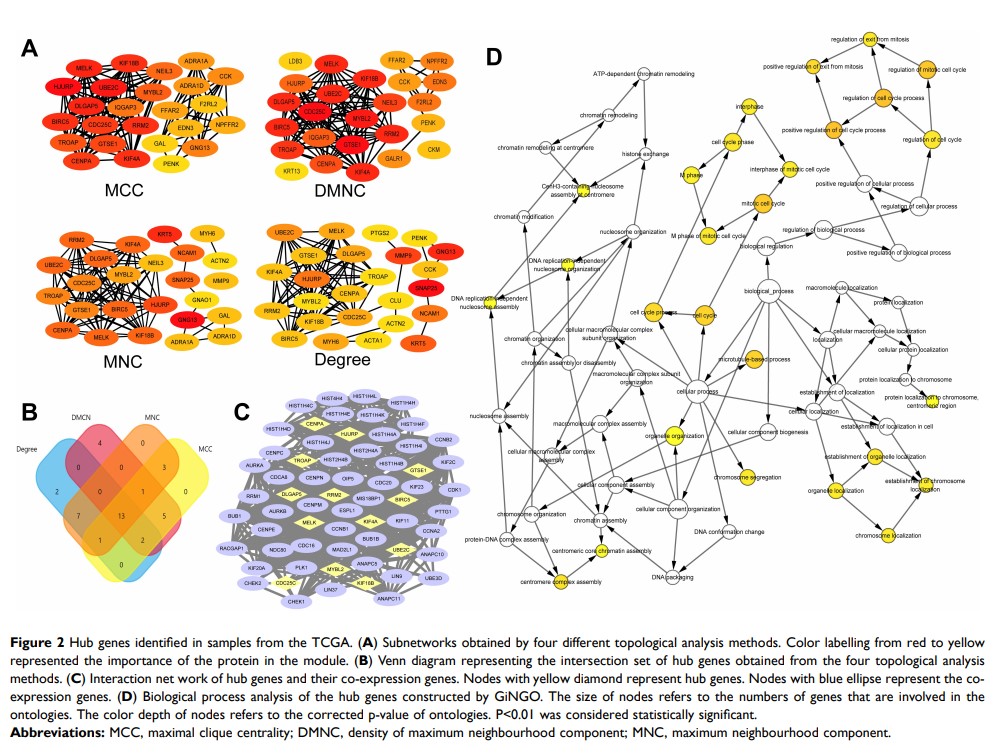
Materials and Methods: The RNA-seq data were downloaded from The Cancer Genome Atlas (TCGA) database to identify the differentially expressed genes (DEGs). The hub genes were screened by building protein–protein interaction (PPI) subnetworks with four topological analysis methods. The overall survival analysis of hub genes was conducted using Kaplan–Meier curves. Furthermore, the bioinformatics results were confirmed in 102 PCa samples collected in our hospital. Gene Set Enrichment Analysis (GSEA) was performed to provide information about the molecular mechanisms underlying PCa.
Results: Among 13 hub genes, the high expression of GTSE1 or KIF18B was associated with worse overall survival according to the TCGA samples. Immunoreactive scores for GTSE1 staining were significantly higher in PCa tissues than in paracancerous tissues (P< 0.01). The overall survival time of patients with high GTSE1 expression was shorter than that of patients with low GTSE1 expression (P=0.015). GSEA demonstrated that high GTSE1 expression was mainly enriched in the cell cycle (P< 0.001), DNA replication (P< 0.001), mismatch repair (P< 0.001), and p53 signaling pathway (P< 0.001).
Conclusion: GTSE1 expression was significantly high in PCa and associated with poor prognosis. GTSE1 may serve as a potential biomarker and therapeutic target in PCa patients.
Keywords: prostate cancer, GTSE1, bioinformatics analysis, prognosis