110932
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

通过生物信息学分析进行头颈鳞状细胞癌潜在生物标志物的鉴定和预后价值分析
Authors Yang B, Chen Z, Huang Y, Han G, Li W
Received 24 February 2017
Accepted for publication 4 April 2017
Published 26 April 2017 Volume 2017:10 Pages 2315—2321
DOI https://doi.org/10.2147/OTT.S135514
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Narasimha Reddy Parine
Peer reviewer comments 2
Editor who approved publication: Dr Yao Dai
Abstract: The purpose of this study was to find disease-associated genes and
potential mechanisms in head and neck squamous cell carcinoma (HNSCC) with
deoxyribonucleic acid microarrays. The gene expression profiles of GSE6791 were
downloaded from the Gene Expression Omnibus database. Differentially expressed
genes (DEGs) were obtained with packages in R language and STRING constructed
protein–protein interaction (PPI) network of the DEGs with combined score
>0.8. Subsequently, module analysis of the PPI network was performed by
Molecular Complex Detection plugin and functions and pathways of the hub gene
in subnetwork were studied. Finally, overall survival analysis of hub genes was
verified in TCGA HNSCC cohort. A total of 811 DEGs were obtained, which were
mainly enriched in the terms related to extracellular matrix (ECM)–receptor
interaction, ECM structural constituent, and ECM organization. A PPI network
was constructed, consisting of 401 nodes and 1,254 edges and 15 hub genes with
high degrees in the network. High expression of 4 genes of the 15 genes was
associated with poor OS of patients in HNSCC, including PSMA7 , ITGA6 , ITGB4 , and APP . Two
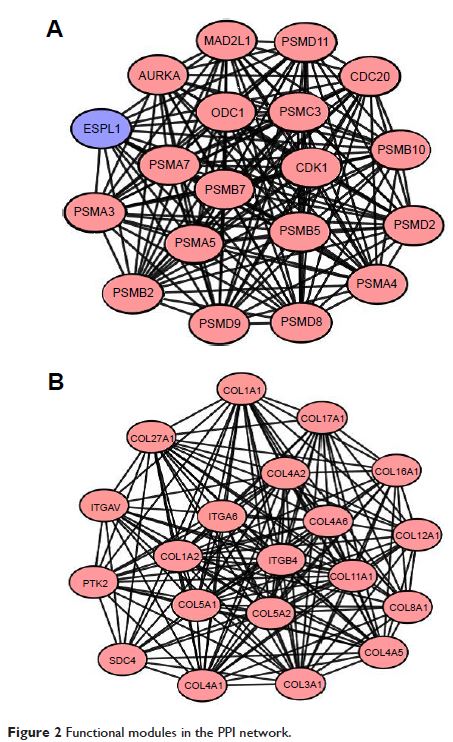
significant modules were detected from the PPI network, and the enriched
functions and pathways included proteasome, ECM organization, and ECM–receptor
interaction. In conclusion, we propose that PSMA7 , ITGA6 , ITGB4 , and APP may be further explored as potential
biomarkers to aid HNSCC diagnosis and treatment.
Keywords: head and neck squamous cell carcinoma,
interaction network, prognostic biomarkers, function and pathway analysis