111314
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

将 COX5B 作为高级别胶质瘤患者的新型生物标志物
Authors Hu T, Xi J
Received 9 April 2017
Accepted for publication 22 September 2017
Published 15 November 2017 Volume 2017:10 Pages 5463—5470
DOI https://doi.org/10.2147/OTT.S139243
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Akshita Wason
Peer reviewer comments 2
Editor who approved publication: Dr Samir Farghaly
Background: Malignant glioma is the second leading cause of cancer-related
death worldwide, and is known to exhibit a high degree of heterogeneity in its
deregulation of different oncogenic pathways. The molecular subclasses of human
glioma are not well known. Thus, it is crucial to identify vital oncogenic
pathways in glioma with significant relationships to patient survival.
Methods: In this study, we devised a bioinformatics strategy to
map patterns of oncogenic pathway activation in glioma, from the Gene
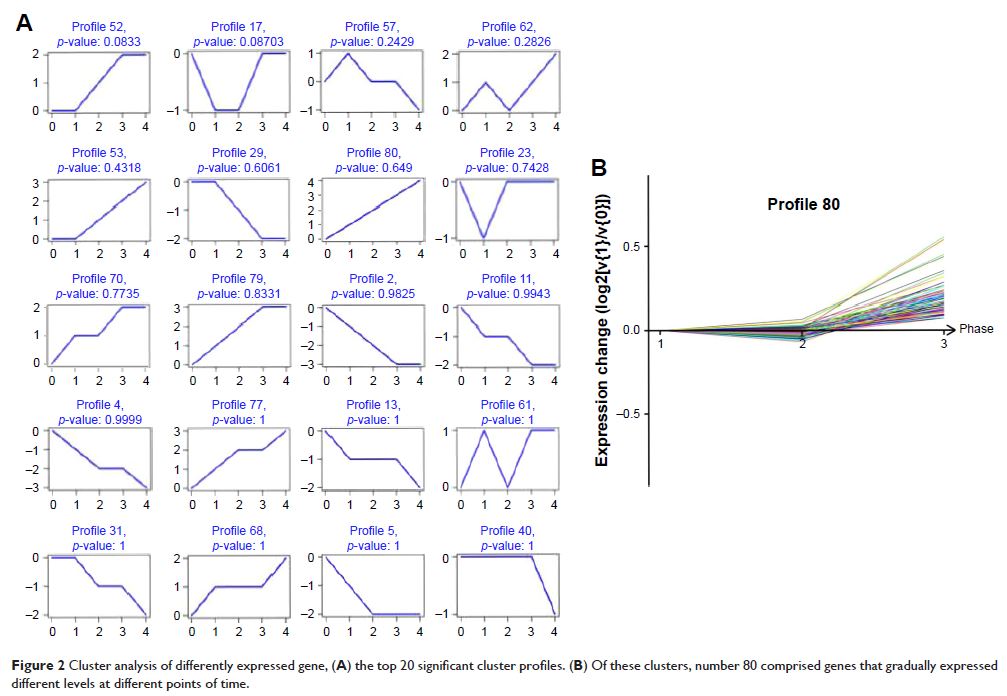
Expression Omnibus (GEO). Bioinformatics analysis revealed that 749 genes were
differentially expressed and classified into different glioma grades.
Results: Using gene expression signatures, we identified three
oncogenic pathways (MAPK signaling pathway, Wnt signaling pathway, and ErbB
signaling pathway) deregulated in the majority of human glioma. Following gene
microarray analysis, the gene expression profile in the differential grade
glioma was further validated by bioinformatic analyses, with coexpression
network construction. Furthermore, we found that cytochrome c oxidase subunit
Vb (COX5B), the terminal enzyme of the electron transport chain, was the
central gene in a coexpression network that transfers electrons from reduced
cytochrome c to oxygen and, in the process, generates an electrochemical
gradient across the mitochondrial inner membrane. The expression level of COX5B
was then detected in 87 glioma tissues as well as adjacent normal tissues using
immunohistochemistry. We found that COX5B was significantly upregulated in 67
of 87 (77.0%) glioma and glioblastoma tissues, compared with adjacent tissue (p <0.01). Furthermore,
statistical analysis showed the COX5B expression level was significantly
associated with clinical stage and lymph node status, while there were no
correlations between COX5B expression and age or tumor size.
Conclusion: These data indicate that COX5B may be implicated in
glioma pathogenesis and as a biomarker for identification of the pathological
grade of glioma.
Keywords: bioinformatics,
COX5B, biomarker, glioblastoma