110932
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

通过综合分析显示在肺腺癌中具有预后价值的关键基因
Authors Song YJ, Tan J, Gao XH, Wang LX
Received 19 March 2018
Accepted for publication 4 August 2018
Published 21 November 2018 Volume 2018:10 Pages 6097—6108
DOI https://doi.org/10.2147/CMAR.S168636
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Amy Norman
Peer reviewer comments 3
Editor who approved publication: Dr Antonella D'Anneo
Background: Lung
cancer is one of the most common malignant tumors. Despite advances in lung
cancer therapies, prognosis of non-small-cell lung cancer is still unfavorable.
The aim of this study was to identify the prognostic value of key genes in lung
tumorigenesis.
Methods: Differentially
expressed genes (DEGs) were screened out by GEO2R from three Gene Expression
Omnibus cohorts. Common DEGs were selected for Kyoto Encyclopedia of Genes and
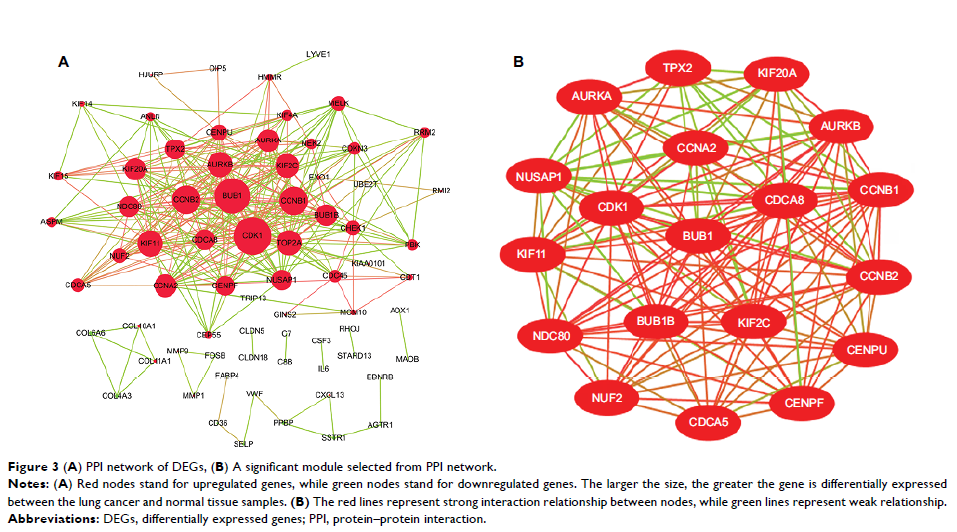
Genomes pathway analysis and Gene Ontology enrichment analysis. Protein–protein
interaction networks were constructed by the STRING database and visualized by
Cytoscape software. Hub genes, filtered from the CytoHubba, were validated
using the Gene Expression Profiling Interactive Analysis database, and their
genomic alterations were identified by performing the cBioportal. Finally,
overall survival analysis of hub genes was performed using Kaplan–Meier
Plotter.
Results: From
three datasets, 169 DEGs (70 upregulated and 99 downregulated) were identified.
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway analyses
showed that upregulated DEGs were significantly enriched in cell cycle, p53
pathway, and extracellular matrix–receptor interactions; the downregulated DEGs
were significantly enriched in PPAR pathway and tyrosine metabolism. The
protein–protein interaction network consisted of 71 nodes and 305 edges, including
49 upregulated and 22 downregulated genes. The hub genes, including AURKB , BUB1B , KIF2C , HMMR , CENPF , and CENPU , were
overexpressed compared with the normal group by Gene Expression Profiling
Interactive Analysis analysis, and associated with reduced overall survival in
lung cancer patients. In the genomic alterations analysis, two hotspot
mutations (S2021C/F and E314K/V) were identified in Pfam protein domains.
Conclusion: DEGs,
including AURKB , BUB1B , KIF2C , HMMR , CENPF , and CENPU , might be
potential biomarkers for the prognosis and treatment of lung adenocarcinoma.
Keywords: lung
adenocarcinoma, prognosis, gene expression profiling, differentially expressed,
bioinformatics analysis