110932
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

CCNA2 作为调节结直肠癌生长和凋亡的新型生物标志物
Authors Gan Y, Li Y, Li T, Shu G, Yin G
Received 11 June 2018
Accepted for publication 1 September 2018
Published 31 October 2018 Volume 2018:10 Pages 5113—5124
DOI https://doi.org/10.2147/CMAR.S176833
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Colin Mak
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Objective: Colorectal cancer (CRC) is considered to be the most prevalent malignant tumors that contribute to high cancer-related mortality. However, the signaling pathways involved in CRC and CRC-driven genes are largely unknown. We seek to discover a novel biomarker in CRC.
Materials and methods: All clinical CRC samples (n=33) were from Xiangya Hospital. We first selected CCNA2 by integrated bioinformatics analysis of four GSE databases. Next, the expression of CCNA2 in tissues and cell lines was verified by quantitative real-time PCR. The effects of CCNA2 on cell growth, proliferation, cell cycle, and apoptosis were examined by in vitro assays.
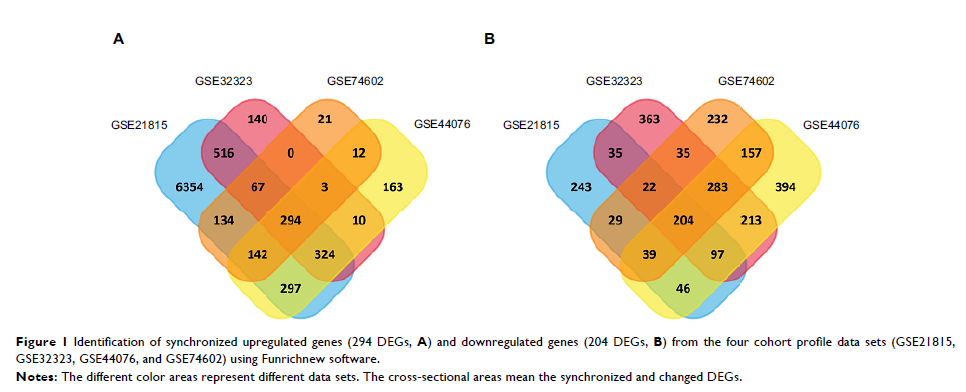
Results: We identified 498 shared DEGs (294 upregulated and 204 downregulated), and the top ten hub genes were selected by integrated analysis. These hub genes were significantly overexpressed in CRC samples and were positively correlated. Our data revealed that the expression of CCNA2 in CRC tissues is higher than that in normal tissues. The CCNA2 knockdown could significantly suppress CRC cell growth by impairing cell cycle progression and inducing cell apoptosis.
Conclusion: CCNA2 , as a novel oncogenic gene, plays a role in regulating cancer cell growth and apoptosis. It could be used as a new biomarker for diagnosis and therapy in CRC.
Keywords: CCNA2, colorectal cancer, bioinformatics analysis, proliferation, cell cycle, apoptosis