110932
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

在癌症基因组图谱数据集中鉴定有淋巴结转移的早期宫颈癌中的差异表达的 miRNA
Authors Chen Q, Zeng X, Huang D, Qiu X
Received 10 August 2018
Accepted for publication 9 November 2018
Published 28 November 2018 Volume 2018:10 Pages 6489—6504
DOI https://doi.org/10.2147/CMAR.S183488
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Colin Mak
Peer reviewer comments 2
Editor who approved publication: Dr Beicheng Sun
Background and aim: Previous
studies have suggested that lymph node metastasis (LNM) in early-stage cervical
cancer (CESC) may affect the prognosis of patients and the outcomes of
subsequent adjuvant therapy. However, research focused on miRNA expression in
early-stage CESC patients with LNM remains limited. Therefore, it is necessary
to identify prognostic miRNAs and determine their molecular mechanisms.
Methods: We
evaluated the differentially expressed genes in early-stage CESC patients with
LNM compared to patients without LNM and evaluated the prognostic significance
of these differentially expressed genes by analyzing a public dataset from The
Cancer Genome Atlas. Potential molecular mechanisms were investigated by gene
ontology, the Kyoto Encyclopedia of Genes and Genomes, and protein–protein
interaction network analyses.
Results: According
to the The Cancer Genome Atlas data, hsa-miR-508, hsa-miR-509-2, and
hsa-miR-526b expression levels were significantly lower in early-stage CESC
patients with LNM than in patients without LNM. A multivariate analysis
suggested that three miRNAs were prognostic factors for CESC (P <0.05). The
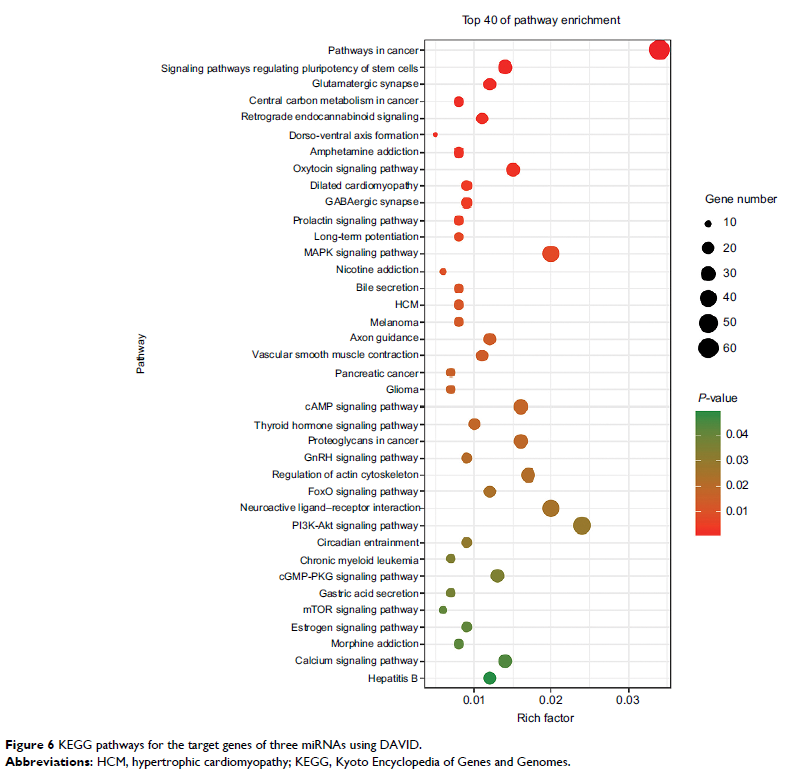
target genes were identified to be involved in the MAPK, cAMP, PI3K/Akt, mTOR,
and estrogen cancer signaling pathways. Protein–protein interaction network
analysis showed that TP53, MMP1, NOTCH1, SMAD4, and NFKB1 were the most
significant hub proteins.
Conclusion: Our
results indicate that hsa-miR-508, hsa-miR-509-2, and hsa-miR-526b may be
potential diagnostic biomarkers for early-stage CESC with LNM, and serve as
prognostic predictors for patients with CESC.
Keywords: lymph
nodes, miRNAs, prognosis, uterine cervical neoplasms