110932
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

循环肿瘤 DNA 的新一代测序用于检测肺癌基因突变:对精准治疗的影响
Authors Lai J, Du B, Wang Y, Wu R, Yu Z
Received 21 May 2018
Accepted for publication 23 August 2018
Published 14 December 2018 Volume 2018:11 Pages 9111—9116
DOI https://doi.org/10.2147/OTT.S174877
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Narasimha Reddy Parine
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Background: Lung cancer
remains a major global health problem, which causes millions of deaths
annually. Because the prognosis is mainly determined by the stage of lung
cancer, precise early diagnosis is of great significance to improve the
survival and prognosis. Circulating tumor DNA (ctDNA) has been recognized as a
sensitive and specific biomarker for the detection of early- and late-stage
lung cancer, and next-generation sequencing (NGS) of ctDNA has been accepted as
a noninvasive tool for early identification and monitoring of cancer mutations.
This study aimed to assess the value of NGS-based ctDNA analysis in detecting
gene mutations in lung cancer patients.
Methods: A total
of 101 subjects with pathological diagnosis of lung cancer were enrolled, and
blood samples were collected. ctDNA samples were prepared and subjected to NGS
assays.
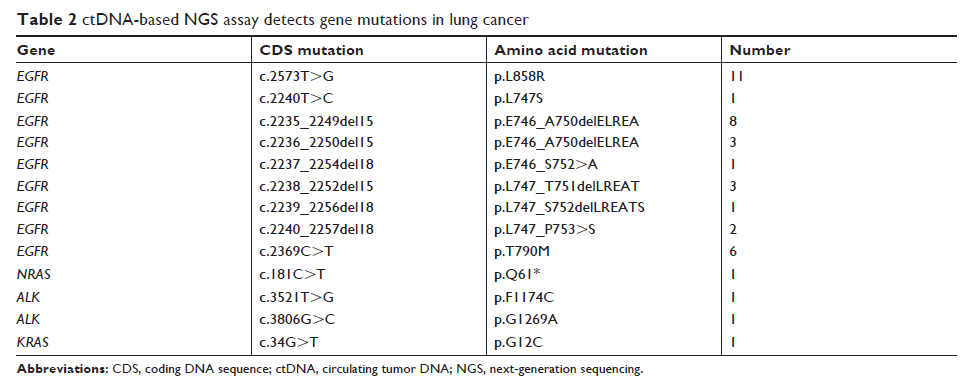
Results: There were 31
cases harboring 40 gene mutations, and EGFR was the most frequently mutated gene
(27.72%). In addition, there were seven cases with double mutations and one
case with triple mutations, with EGFR p.T790M mutation exhibiting the highest
frequency.
Conclusion: Our findings
demonstrate that NGS of ctDNA is effective in detecting gene mutations in lung
cancer patients, and may be used as a liquid biopsy for lung cancer, which
facilitates the development of precision treatment regimens for lung cancer.
Keywords: lung cancer,
next-generation sequencing, circulating tumor DNA, ctDNA, gene mutation