110932
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

6 mRNA 预后模型用于预测头颈部鳞状细胞癌存活
Authors Tian S, Meng G, Zhang W
Received 1 September 2018
Accepted for publication 15 November 2018
Published 20 December 2018 Volume 2019:11 Pages 131—142
DOI https://doi.org/10.2147/CMAR.S185875
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Andrew Yee
Peer reviewer comments 4
Editor who approved publication: Dr Ahmet Emre Eskazan
Background: Transcriptional
dysregulation is one of the most important features of cancer genesis and
progression. Applying gene expression dysregulation information to predict the
development of cancers is useful for cancer diagnosis. However, previous
studies mainly focused on the relationship between a single gene and cancer.
Prognostic prediction using combined gene models remains limited.
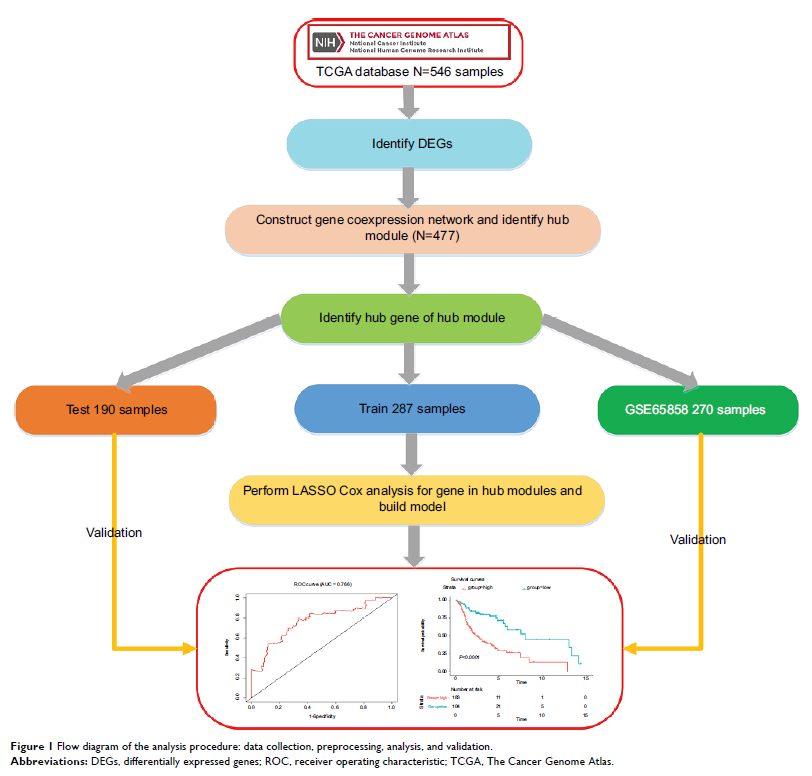
Materials and methods: Gene
expression profiles were downloaded from The Cancer Genome Atlas and the data
sets were randomly divided into training data sets and test data sets. A
six-gene signature associated with head and neck squamous cell carcinoma
(HNSCC) and overall survival (OS) was identified according to a training cohort
by using weighted gene correlation network analysis and least absolute
shrinkage and selection operator Cox regression. The test data set and gene
expression omnibus (GEO) data set were used to validate this signature.
Results: We
identified six candidate genes, namely, FOXL2NB, PCOLCE2, SPINK6, ULBP2,
KCNJ18, and RFPL1, and, using a six-gene model, predicted the risk of death of
head and neck squamous cell carcinoma in The Cancer Genome Atlas. At a selected
cutoff, patients were clustered into low- and high-risk groups. The OS curves
of the two groups of patients had significant differences, and the
time-dependent receiver operating characteristics of OS, disease-specific
survival (DSS), and progression-free survival (PFS) were as high as 0.766,
0.731, and 0.623, respectively. Then, the test data set and the GEO data set
were used to evaluate our model, and we found that the OS time in the high-risk
group was significantly shorter than in the low-risk group in both data sets,
and the receiver operating characteristics of test data set were 0.669, 0.675,
and 0.614, respectively. Furthermore, univariate and multivariate Cox
regression analyses showed that the risk score was independent of
clinicopathological features.
Conclusion: The
six-gene model could predict the OS of HNSCC patients and improve therapeutic
decision-making.
Keywords: gene
expression dysregulation, TCGA, six-gene model, OS