110932
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

通过加权基因共表达网络分析、Cox 回归和 L1-LASSO 处罚对胶质母细胞瘤中的长非编码 RNA 进行分析,用于预后预测
Authors Liang R, Zhi YQ, Zheng G, Zhang B, Zhu H, Wang M
Received 22 April 2018
Accepted for publication 4 September 2018
Published 21 December 2018 Volume 2019:12 Pages 157—168
DOI https://doi.org/10.2147/OTT.S171957
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Andrew Yee
Peer reviewer comments 3
Editor who approved publication: Dr William Cho
Purpose: This
study focused on identification of long non-coding RNAs (lncRNAs) for prognosis
prediction of glioblastoma (GBM) through weighted gene co-expression network
analysis (WGCNA) and L1-penalized least absolute shrinkage and selection
operator (LASSO) Cox proportional hazards (PH) model.
Materials and methods: WGCNA was
performed based on RNA expression profiles of GBM from Chinese Glioma Genome
Atlas (CGGA), National Center for Biotechnology Information (NCBI) Gene
Expression Omnibus (GEO), and the European Bioinformatics Institute
ArrayExpress for the identification of GBM-related modules. Subsequently,
prognostic lncRNAs were determined using LASSO Cox PH model, followed by
constructing a risk scoring model based on these lncRNAs. The risk score was
used to divide patients into high- and low-risk groups. Difference in survival
between groups was analyzed using Kaplan–Meier survival analysis. IncRNA-mRNA
networks were built for the prognostic lncRNAs, followed by pathway enrichment
analysis for these networks.
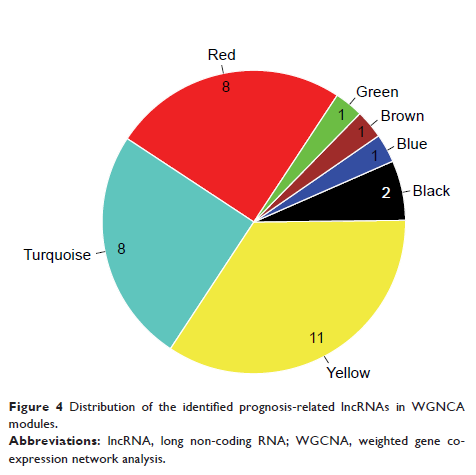
Results: This
study identified eight preserved GBM-related modules, including 188 lncRNAs.
Consequently, C20orf166-AS1, LINC00645, LBX2-AS1, LINC00565, LINC00641, and
PRRT3-AS1 were identified by LASSO Cox PH model. A risk scoring model based on
the lncRNAs was constructed that could divide patients into different risk
groups with significantly different survival rates. Prognostic value of this
six-lncRNA signature was validated in two independent sets. C20orf166-AS1 was
associated with antigen processing and presentation and cell adhesion molecule
pathways, involving nine common genes. LBX2-AS1, LINC00641, PRRT3-AS1, and
LINC00565 were related to focal adhesion, extracellular matrix receptor
interaction, and mitogen-activated protein kinase signaling pathways, which
shared 12 common genes.
Conclusion: This prognostic
six-lncRNA signature may improve prognosis prediction of GBM. This study
reveals many pathways and genes involved in the mechanisms behind these
lncRNAs.
Keywords: lncRNA, risk
score, WGCNA, network, pathway