110932
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

基于生物信息学分析鉴定中枢基因和结肠癌的结果
Authors Yang W, Ma J, Zhou W, Li Z, Zhou X, Cao B, Zhang Y, Liu J, Yang Z, Zhang H, Zhao Q, Hong L, Fan D
Received 6 May 2018
Accepted for publication 29 October 2018
Published 27 December 2018 Volume 2019:11 Pages 323—338
DOI https://doi.org/10.2147/CMAR.S173240
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Colin Mak
Peer reviewer comments 4
Editor who approved publication: Dr Beicheng Sun
Background: Colon
cancer is one of the leading malignant neoplasms worldwide. Until now, the
concrete mechanisms of colonic cancerogenesis are largely unknown;
identification of driven genes and pathways is, therefore, of great importance
for monitoring and conquering this disease. This study aims to explore the
potential biomarkers and therapeutic targets for colon cancer treatment.
Methods: The gene
expression profile of GSE44076 from Gene Expression Omnibus database, including
98 primary colon cancers and 98 normal distant colon mucosa, was deeply
analyzed. GEO2R tool was used to screen the differentially expressed genes
(DEGs) between colon cancer tissues and normal samples. Gene Ontology analysis
and Kyoto Encyclopedia of Genes and Genomes pathway analysis were performed for
screening DEGs using Database for Annotation, Visualization and Integrated
Discovery database and Panther database. Moreover, Search Tool for the
Retrieval of Interacting Genes, Cytoscape software, and Molecular Complex
Detection plug-in were used to visualize the protein–protein interaction of
these DEGs.
Results: A total
of 497 DEGs were obtained, including 129 upregulated genes mainly enriched in
Hippo signaling pathway, Wnt signaling pathway, and cytokine–cytokine receptor
interaction and 368 downregulated genes enriched in retinol metabolism, steroid
hormone biosynthesis, drug metabolism, and chemical carcinogenesis. Using
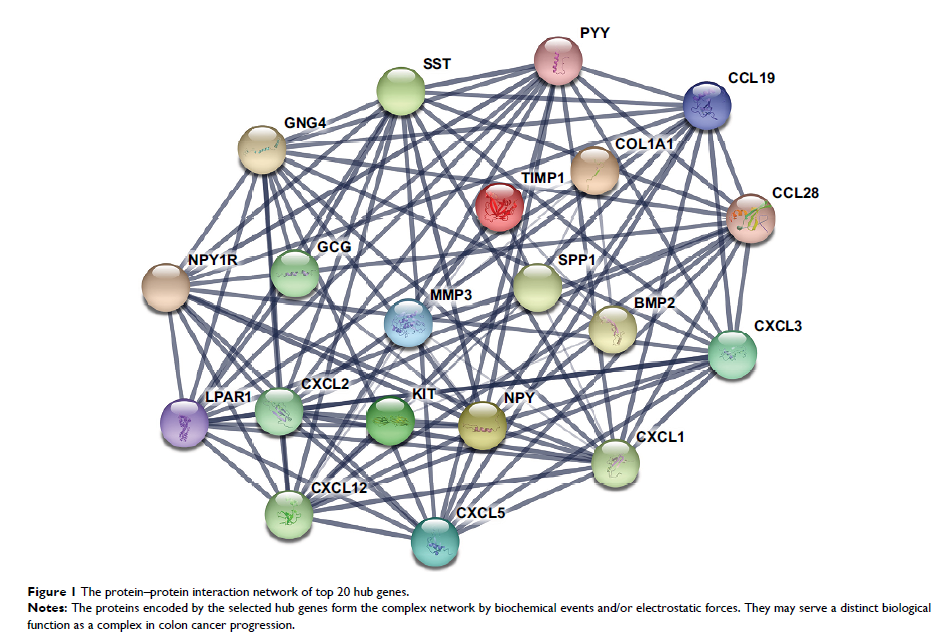
Molecular Complex Detection software, three important modules were selected
from the protein–protein interaction network. Moreover, 20 hub genes with high
degree of connectivity were selected, including COL1A1, CXCL5, GNG4, TIMP1, and
so on. The Kaplan–Meier analysis for overall survival and correlation analysis
were applied among the hub genes.
Conclusion: Taken
together, DEGs, especially the hub genes such as COL1A1, might be the driven
genes in colon cancer progression. More importantly, they might be the novel
biomarkers for diagnosis and guiding therapeutic strategies of colon cancer.
Keywords: colon
cancer, protein–protein interaction, bioinformatics analysis, diagnosis,
prognosis