110932
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

鉴定可能涉及激素非依赖性和米托蒽醌抗性前列腺癌的关键基因和特定通路
Authors Zhu S, Jiang LL, Wang LY, Wang LL, Zhang C, Ma Y, Huang T
Received 7 July 2018
Accepted for publication 16 November 2018
Published 3 January 2019 Volume 2019:11 Pages 419—430
DOI https://doi.org/10.2147/CMAR.S179467
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Amy Norman
Peer reviewer comments 3
Editor who approved publication: Dr Rituraj Purohit
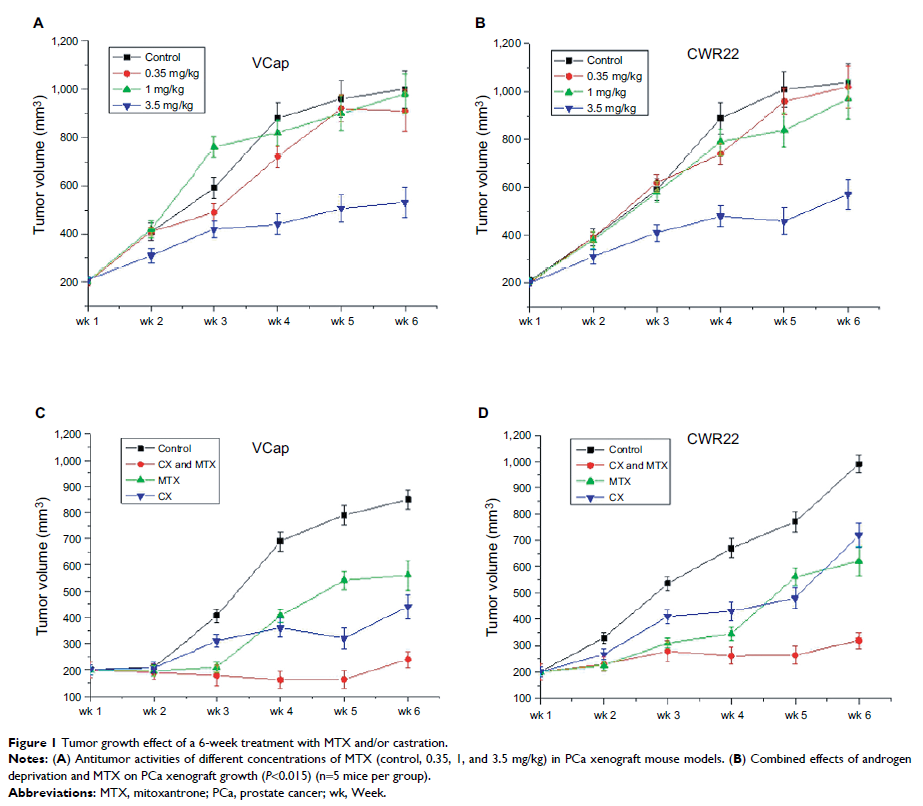
Background: Resistance
to mitoxantrone (MTX), an anthracenedione antineoplastic agent used in advanced
and metastatic androgen-refractory prostate cancer (PCa), seriously limits
therapeutic success.
Methods: Xenografts from
two human PCa cell lines (VCaP and CWR22) were established in male severe
combined immunodeficiency mice, and MTX was administered, with or without
concurrent castration, three times a week until tumors relapsed. Microarray
technology was used to screen for differentially expressed genes (DEGs) in
androgen-independent, MTX-resistant PCa xenografts. Gene expression profiles of
MTX-treatment xenografts and their respective parental cell lines were
performed using an Agilent whole human genome oligonucleotide microarray and
analyzed using Ingenuity Pathway Analysis software.
Results: A total of 636
genes were differentially expressed (fold change ≥1.5; P <0.05) in
MTX-resistant castration-resistant prostate cancer (CRPC) xenografts. Of these,
18 were selected to be validated and showed that most of these genes exhibited
a transcriptional profile similar to that seen in the microarray
(Pearson’s r =0.87).
Western blotting conducted with a subset of genes deregulated in MTX-resistant
CRPC tumors was shown through network analysis to be involved in androgen
synthesis, drug efflux, ATP synthesis, and vascularization.
Conclusion: The present
data provide insight into the genetic alterations underlying MTX resistance in
androgen-independent PCa and highlight potential targets to improve therapeutic
outcomes.
Keywords: castration-resistant
prostate cancer, gene expression profiling, drug resistance, differentially
expressed genes