110932
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

将 TCGA 和 GEO 数据库的数据与逆转录定量 PCR 验证相结合以鉴定肺癌中的基因预后标志物
Authors Liu X, Wang J, Chen M, Liu S, Yu X, Wen F
Received 17 August 2018
Accepted for publication 5 December 2018
Published 21 January 2019 Volume 2019:12 Pages 709—720
DOI https://doi.org/10.2147/OTT.S183944
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Justinn Cochran
Peer reviewer comments 2
Editor who approved publication: Dr Leo Jen-Liang Su
Background: The aim
of this study was to predict and explore the possible mechanism and clinical
value of genetic markers in the development of lung cancer with a combined
database to screen the prognostic genes of lung cancer.
Materials and methods: Common
differential genes in two gene expression chips (GSE3268 and GSE10072 datasets)
were investigated by collecting and calculating from Gene Expression Omnibus
and The Cancer Genome Atlas databases using R language. Five markers of gene
composition (ribonucleotide reductase regulatory subunit M2 [RRM2], trophoblast
glycoprotein [TPBG], transmembrane protease serine 4[TMPRFF4], chloride
intracellular channel 3 [CLIC3], and WNT inhibitory factor-1 [WIF1]) were found
by the stepwise Cox regression function when we further screened combinations
of gene models, which were more meaningful for prognosis. By analyzing the
correlation between gene markers and clinicopathological parameters of lung
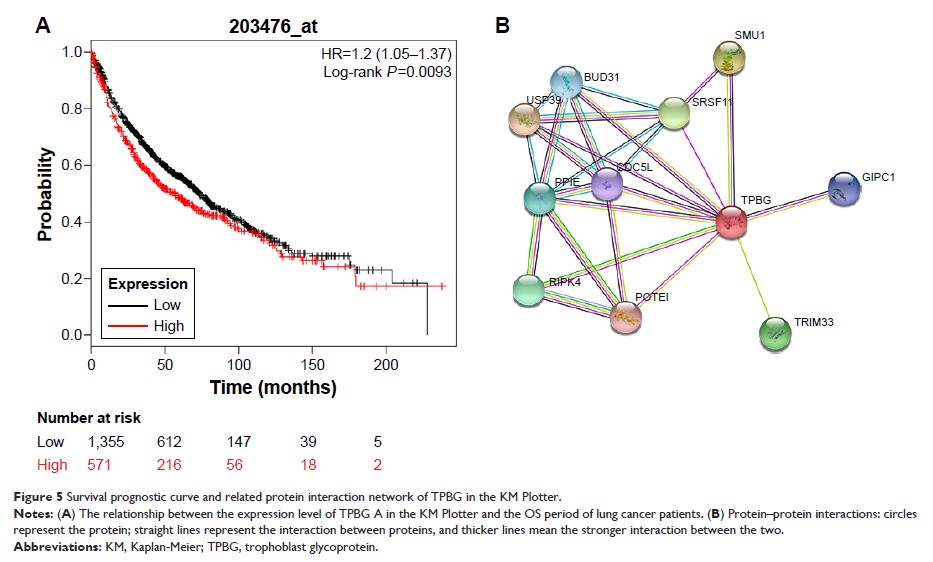
cancer and its effect on prognosis, the TPBG gene was selected to analyze
differential expression, its possible pathways and functions were predicted
using gene set enrichment analysis (GSEA), and its protein interaction network
was constructed using the Search Tool for the Retrieval of Interacting
Genes/Proteins (STRING) database; then, quantitative PCR and the Oncomine
database were used to verify the expression differences of TPBG in lung cancer
cells and tissues.
Results: The
expression levels of five genetic markers were correlated with survival
prognosis, and the total survival time of the patients with high expression of
the genetic markers was shorter than those with low expression (P <0.001). GSEA
showed that these high-expression samples enriched the gene sets of cell
adhesion, cytokine receptor interaction pathway, extracellular matrix receptor
pathway, adhesion pathway, skeleton protein regulation, cancer pathway and TGF-β
pathway.
Conclusion: The high
expression of five gene constituent markers is a poor prognostic factor in lung
cancer and may serve as an effective biomarker for predicting metastasis and
prognosis of patients with lung cancer.
Keywords: lung
cancer, prognostic genes, GEO, TCGA, bioinformatics analysis, TPBG