110932
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

活动性和潜伏性结核的血液转录特征
Authors Deng M, Lv XD, Fang ZX, Xie XS, Chen WY
Received 20 August 2018
Accepted for publication 27 December 2018
Published 30 January 2019 Volume 2019:12 Pages 321—328
DOI https://doi.org/10.2147/IDR.S184640
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Amy Norman
Peer reviewer comments 2
Editor who approved publication: Dr Eric Nulens
Background: Although
the incidence of tuberculosis (TB) has dropped substantially, it still is a
serious threat to human health. And in recent years, the emergence of resistant
bacilli and inadequate disease control and prevention has led to a significant
rise in the global TB epidemic. It is known that the cause of TB is Mycobacterium tuberculosis infection.
But it is not clear why some infected patients are active while others are
latent.
Methods: We
analyzed the blood gene expression profiles of 69 latent TB patients and 54
active pulmonary TB patients from GEO (Transcript Expression Omnibus) database.
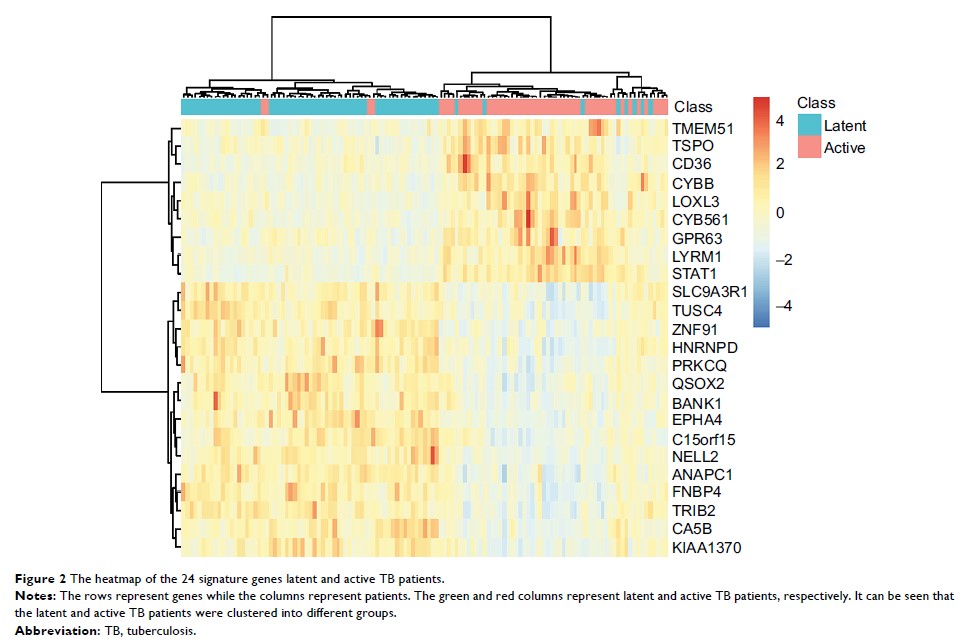
Results: By
applying minimal redundancy maximal relevance and incremental feature
selection, we identified 24 signature genes which can predict the TB
activation. The support vector machine predictor based on these 24 genes had a
sensitivity of 0.907, specificity of 0.913, and accuracy of 0.911,
respectively. Although they need to be validated in a large independent
dataset, the biological analysis of these 24 genes showed great promise.
Conclusion: We found
that cytokine production was a key process during TB activation and genes like
CYBB, TSPO, CD36, and STAT1 worth further investigation.
Keywords: tuberculosis,
blood gene expression, support vector machine, minimal redundancy maximal
relevance, incremental feature selection