110932
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

使用基于核磁共振的血浆代谢组学对脑卒中后抑郁症进行客观诊断
Authors Hu Z, Fan S, Liu M, Zhong J, Cao D, Zheng P, Wang Y, Wei Y, Fang L, Xie P
Received 26 October 2018
Accepted for publication 3 March 2019
Published 10 April 2019 Volume 2019:15 Pages 867—881
DOI https://doi.org/10.2147/NDT.S192307
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Andrew Yee
Peer reviewer comments 2
Editor who approved publication: Dr Jun Chen
Background: Post-stroke
depression (PSD) is a frequent and serious complication of stroke. However, the
underlying molecular basis of PSD remains largely unknown, and no empirical
laboratory tests were available to diagnose this disorder.
Materials and methods: A proton
nuclear magnetic resonance (1H NMR)-based metabonomic approach was employed
to profile plasma samples from 32 PSD, 35 stroke patients and 35 healthy
comparison subjects (the training set) in order to identify metabolite
biomarkers for PSD. Then, 10 PSD, 11 stroke patients and 11 healthy comparison
subjects (test set) were used to validate the diagnostic performance of these
biomarkers.
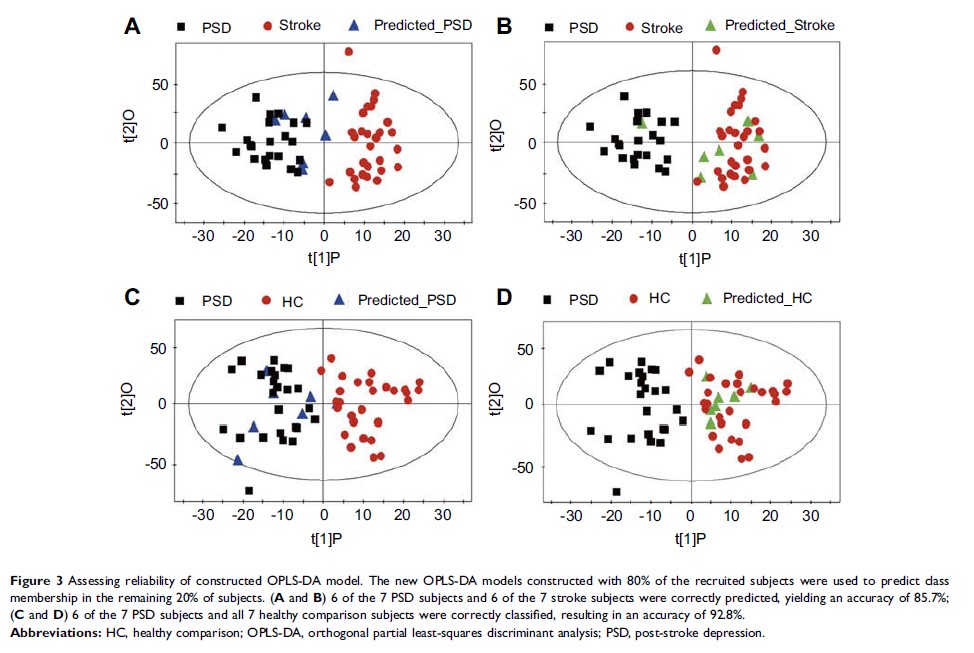
Results: The
multivariate statistical analysis demonstrated that PSD group was significantly
distinguishable from non-PSD groups (non-depression stroke patients and healthy
comparison group). Five plasma metabolites (phenylalanine, tyrosine,
1-methylhistidine, 3-methylhistidine and LDL CH3-(CH2)n-) were identified
responsible for distinguishing PSD from non-PSD subjects. These metabolites
were mainly involved in neurotransmitter metabolism and oxidative stress. The
biomarker panel composing of these metabolites was capable of distinguishing
test samples with a sensitivity of 100.0% and a specificity of 95.5%.
Conclusion: Our
findings suggest that plasma disturbances of neurotransmitter levels and
oxidative stress were implicated in the onset of PSD; these disturbed metabolites
biomarkers facilitate to the development of diagnostic tool for PSD.
Keywords: post-stroke
depression, stroke, metabonomics, diagnosis, NMR