110932
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

通过生物信息学分析鉴定精原细胞瘤中的关键基因和途径
Authors Chen YH, Lin TT, Wu YP, Li XD, Chen SH, Xue XY, Wei Y, Zheng QS, Huang JB, Xu N
Received 21 December 2018
Accepted for publication 4 April 2019
Published 14 May 2019 Volume 2019:12 Pages 3683—3693
DOI https://doi.org/10.2147/OTT.S199115
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Ms Shreya Arora
Peer reviewer comments 2
Editor who approved publication: Dr Sanjay Singh
Background: Seminoma
accounts for the most part of cases of testicular germ cell tumor, which is the
most common malignancy among males between ages 15 and 44 years. Understanding
the molecular mechanism of tumorigenesis is important for better clinical
diagnosis and treatment.
Purpose: We
performed bioinformatics analysis to better understand seminoma at the genetic
level and to explore potential candidate genes or molecules for diagnosis,
treatment, and prognosis.
Methods: A
gene expression profile (GSE8607), containing 40 seminoma samples and three
healthy testes samples, was analyzed to identify differentially expressed genes
(DEGs) associated with the occurrence of seminoma. Functional annotation was
then performed using gene ontology and Kyoto Encyclopedia of Genes and Genomes
pathway enrichment analyses. Cytoscape with Search Tool for the Retrieval of
Interacting Genes was used to construct a protein–protein interaction (PPI)
network of the DEGs and select hub genes. Moreover, validation of expression
level and Kaplan–Meier analysis for overall survival were conducted to those
hub genes.
Results: A
total of 1,636 DEGs were identified between seminoma and healthy samples, including
701 up-regulated in seminoma that were enriched in the regulation of immune
responses, defense responses, receptor activity, and signal transducer
activity; 935 were down-regulated in seminoma and were associated with
reproductive processes, kinase activity, and carbohydrate derivative binding.
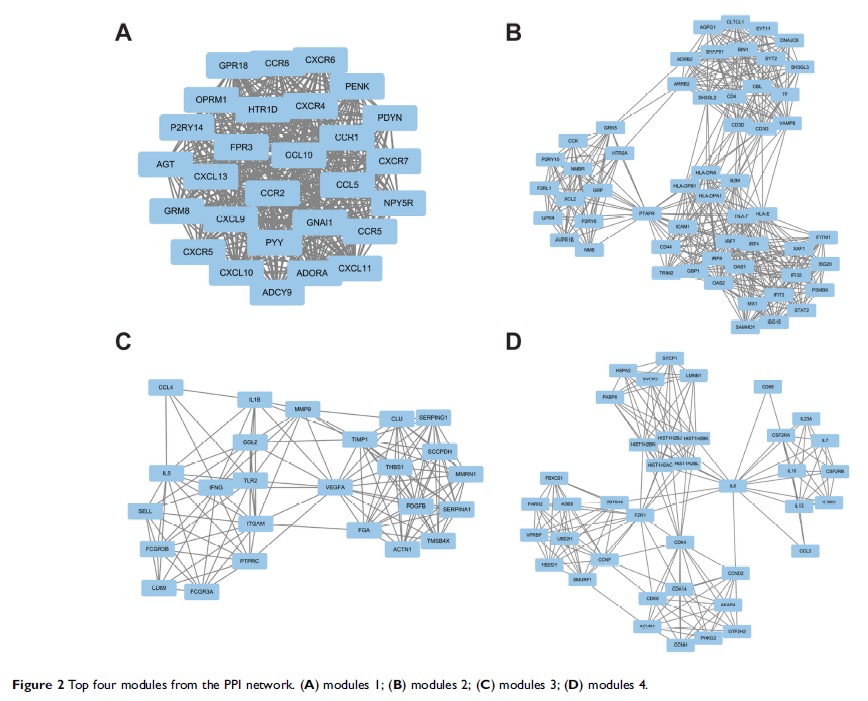
Five hub genes were selected from the PPI network according to the degree of
connectivity: IL6 , VEGFA , IL10 , CCR5 , and CXCR4 . Among them,
high expression levels of CCR5 and CXCR4 were associated with poor prognosis for
seminoma patients. Four modules selected from the PPI network revealed that
seminoma was connected with the Janus kinase-signal transducers and activators
of transcription signaling pathway, chemokine signaling pathway, endocytosis,
and cytokine–cytokine receptor interaction.
Conclusion: These identified DEGs and hub genes facilitate our knowledge of the
underlying molecular mechanism of seminoma and have the potential to be used as
diagnostic biomarkers or therapeutic targets for seminoma.
Keywords: seminoma,
bioinformatics analysis, DEGs, key genes, pathways