110932
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

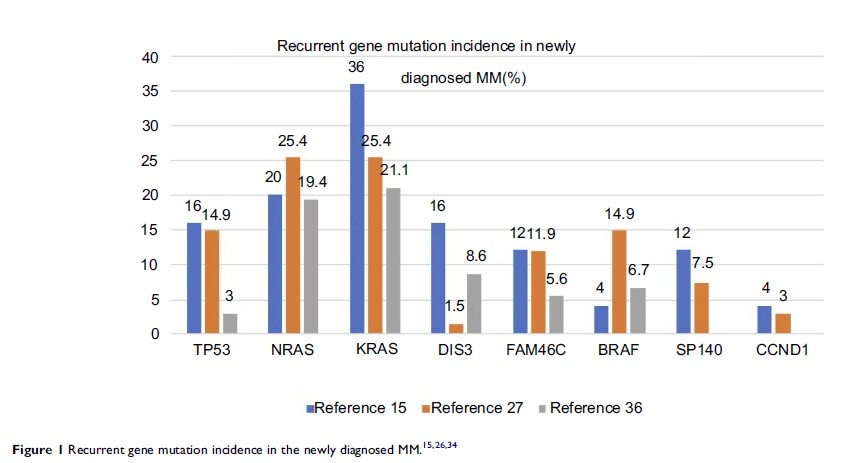
鉴定多发性骨髓瘤基因突变的进展
Authors Hu Y, Chen W, Wang J
Received 19 February 2019
Accepted for publication 30 April 2019
Published 24 May 2019 Volume 2019:12 Pages 4075—4080
DOI https://doi.org/10.2147/OTT.S205922
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Ms Aruna Narula
Peer reviewer comments 2
Editor who approved publication: Dr Takuya Aoki
Abstract: Sequencing
studies have been used to determine a spectrum of multiple myeloma (MM)
mutations. Mutation of certain genes, including KRAS , NRAS , TP53 , FAM46C , DIS3 and BRAF , have a high
recurrence rate and may play important roles in the pathogenesis, progression
and prognosis of MM. Mutations in DIS3 , which encodes a highly conserved RNA
exonuclease, lead to loss of function. The expression of FAM46C is
highly correlated with the expression of ribosomal protein, but the exact function
of FAM46C mutation
is unclear. There are mutants of IRF4 , which is considered an MM survival factor.
Mutations in the gene coding for the DNA damage-binding protein (DDB1 ) may affect
interactions with CUL4A , which is part of the cereblon (CRBN ) ubiquitin
ligase complex. IRF4 is part of the complex, which binds to DNA. These
findings might explain the resistance to immunomodulatory. TP53 deletion
or mutation is often present in B-cell malignancies and is associated with low
response rates. Myeloma pathogenic mutations in ATM have been
found in adult lymphatic tumors. XBP1 and PSMB5 mutations may be related to bortezomib
resistance. Multiple gene mutations (KRAS , NRAS and BRAF ) involved in
the same pathway were found a single patient. Identification of driver gene
mutations has brought great hope to the field of individualized, targeted
medicine for MM.
Keywords: multiple
myeloma, cytogenetic abnormalities, gene mutations