110932
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

鉴定与尤文氏肉瘤患者转移和预后不良有关的关键基因和途径
Authors Li G, Zhang P, Zhang W, Lei Z, He J, Meng J, Di T, Yan W
Received 23 November 2018
Accepted for publication 27 February 2019
Published 27 May 2019 Volume 2019:12 Pages 4153—4165
DOI https://doi.org/10.2147/OTT.S195675
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Colin Mak
Peer reviewer comments 3
Editor who approved publication: Dr Jianmin Xu
Background: Ewing
sarcoma (ES) is the second commonest primary malignant bone neoplasm.
Metastatic status at diagnosis strongly predicted poor prognosis of Ewing
sarcoma patients. Yet little was known about the underlying mechanism of ES
metastasis.
Purpose:This study
intended to identify the relationship between key genes/pathways and
metastasis/poor prognosis in Ewing’s sarcoma patients by using bioinformatic
method.
Methods: In this study,
multi-center sequencing data were obtained from the GEO database, including
gene and miRNA expression profile and prognosis information of ES patients.
Differentially expressed genes (DEGs) were identified between primary and
metastasis ES samples by the GEO2R online tool. Gene ontology (Go) and Kyoto
encyclopedia of genes and genomes (KEGG) pathway enrichment analyses of DEGs
were performed. And PPI network analyses were conducted. The ES patient’s
prognostic information was employed for survival analysis, and the potential
relationship between miRNAs and key genes was analyzed.
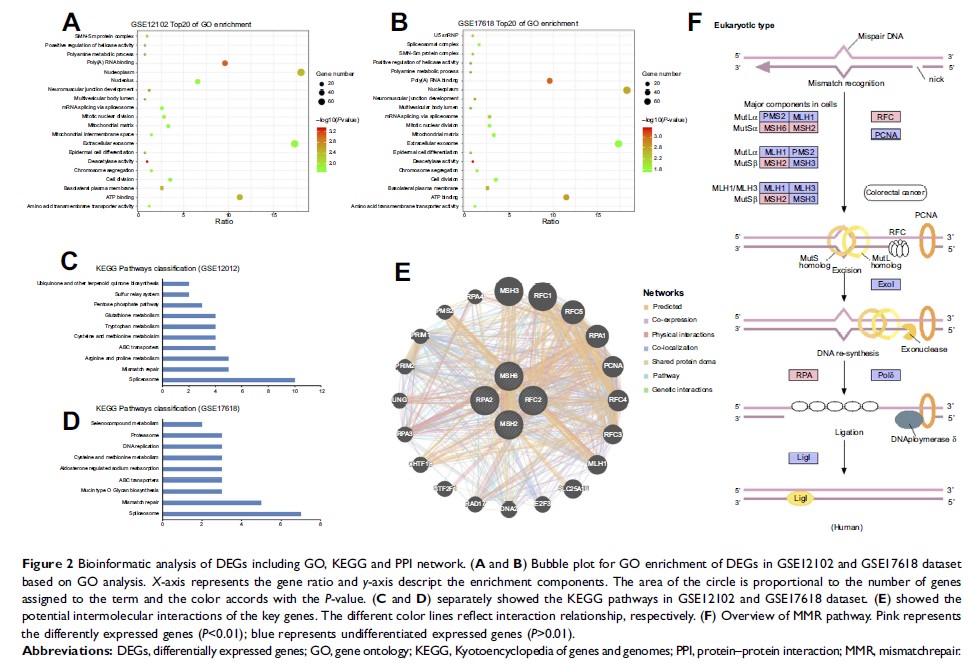
Results: The
results showed that a total of 298 and 428 DEGs were screened out in metastasis
samples based on GSE17618 and GSE12102 dataset compared to primary samples
respectively. The most significantly enriched KEGG pathway was the mismatch
repair (MMR) pathway. MSH2, MSH6, RPA2, and RFC2 that belong to the MMR pathway
were identified as key genes. Moreover, the expression of key genes was
increased in metastasis samples compared with primary ones and was associated
with poor event-free and overall survival of ES patients. The negative
correlation of the expression level of the key genes with patients prognosis
also supported by TCGA sarcoma database. Furthermore, knockdown of EWSR/FLI1
fusion in ES cell line A673 down-regulates the expression of the 4 key genes
was revealed by GDS4962.
Conclusion: In
conclusion, the present study indicated that the key genes promote our
understanding of the molecular mechanisms underlying the development of ES
metastasis, and might be used as molecular targets and diagnostic biomarkers
for the treatment of ES.
Keywords: genes,
miRNAs, Ewing’s sarcoma, metastasis, bioinformatic analysis