110932
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

通过综合生物信息学分析鉴定与宫颈癌的诊断和预后相关的候选生物标志物
Authors Dai F, Chen G, Wang Y, Zhang L, Long Y, Yuan M, Yang D, Liu S, Cheng Y, Zhang L
Received 27 December 2018
Accepted for publication 15 April 2019
Published 12 June 2019 Volume 2019:12 Pages 4517—4532
DOI https://doi.org/10.2147/OTT.S199615
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Ms Aruna Narula
Peer reviewer comments 2
Editor who approved publication: Dr Takuya Aoki
Background: Cervical carcinoma is one of the most common malignant gynecological tumors and is associated with high rates of morbidity and mortality. Early diagnosis and early treatment can reduce the mortality rate of cervical cancer. However, there is still no specific biomarkers for the diagnosis and detection of cervical cancer prognosis. Therefore, it is greatly urgent in searching biomarkers correlated with the diagnosis and prognosis of cervical cancer.
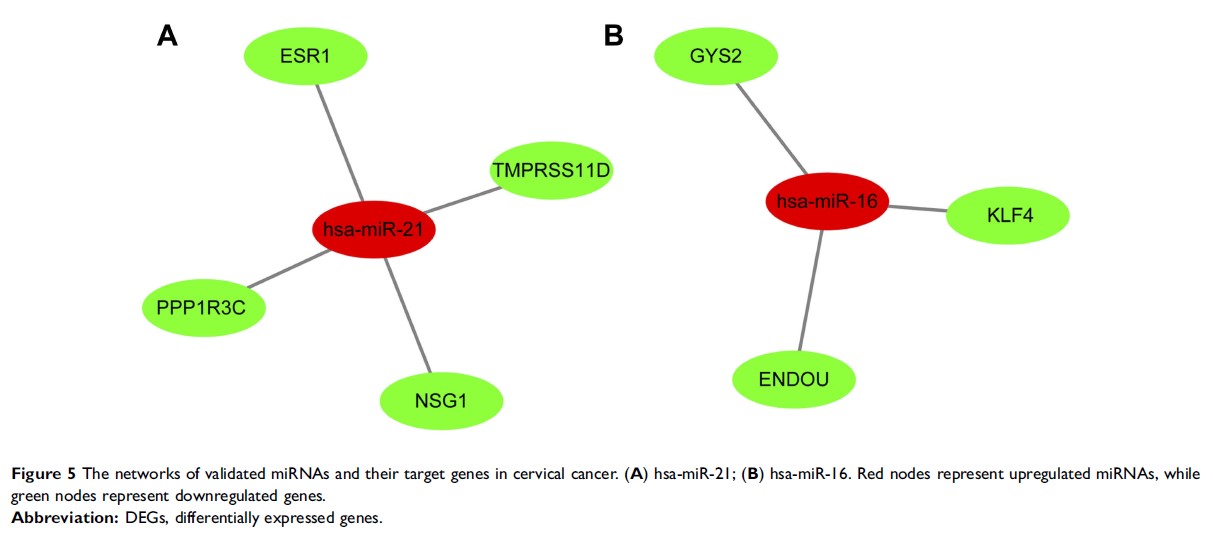
Results: The mRNA and microRNA expression profile datasets (GSE7803, GSE9750, GSE63514, and GSE30656) were downloaded from the Gene Expression Omnibus database (GEO). The three microarray datasets were integrated to one via integrated bioinformatics. Differentially expressed genes (DEGs) and microRNAs (DEMs) were obtained by R software. The protein–protein interaction (PPI) networks of the DEGs were performed from the STRING database and further visualized by Cytoscape software. A total of 83 DEGs and 14 DEMs were screened from the microarray expression profile datasets. The miRNAs validated to be associated with cervical cancer were obtained using HMDD online website and the target genes of DEMs were identified using the miRWalk2.0 online database. ESR1 , PPP1R3C , NSG1 , and TMPRSS11D were the gene targets of hsa-miR-21; the targets of hsa-miR-16 were GYS2 , ENDOU , and KLF4 . These targets were all downregulated in cervical cancer. Finally, we verified the expression of those targets in cervical tissues from TCGA and GTEx databases and analyzed their relationship with survival of cervical cancer patients. In the end, the expression of key genes in cervical cancer tissues was verified via experiment method, we found KLF4 and ESR1 were downregulated in tumor tissues.
Conclusion: This study indicates that KLF4 and ESR1 are downregulated by the upregulated miR21 and miRNA16 in cervical cancer, respectively, using bioinformatics analysis, and the lower expression of KLF4 and ESR1 is closely related to the poor prognosis. They might be of clinical significance for the diagnosis and prognosis of cervical cancer, and provide effective targets for the treatment of cervical cancer.
Keywords: cervical cancer, integrated bioinformatics, differentially expressed genes, differentially expressed miRNAs, biomarkers