111314
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

OSacc:基于基因表达的肾上腺皮质癌生存分析网络工具
Authors Xie L, Wang Q, Nan F, Ge L, Dang Y, Sun X, Li N, Dong H, Han Y, Zhang G, Zhu W, Guo X
Received 14 May 2019
Accepted for publication 2 October 2019
Published 24 October 2019 Volume 2019:11 Pages 9145—9152
DOI https://doi.org/10.2147/CMAR.S215586
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Melinda Thomas
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev Srivastava
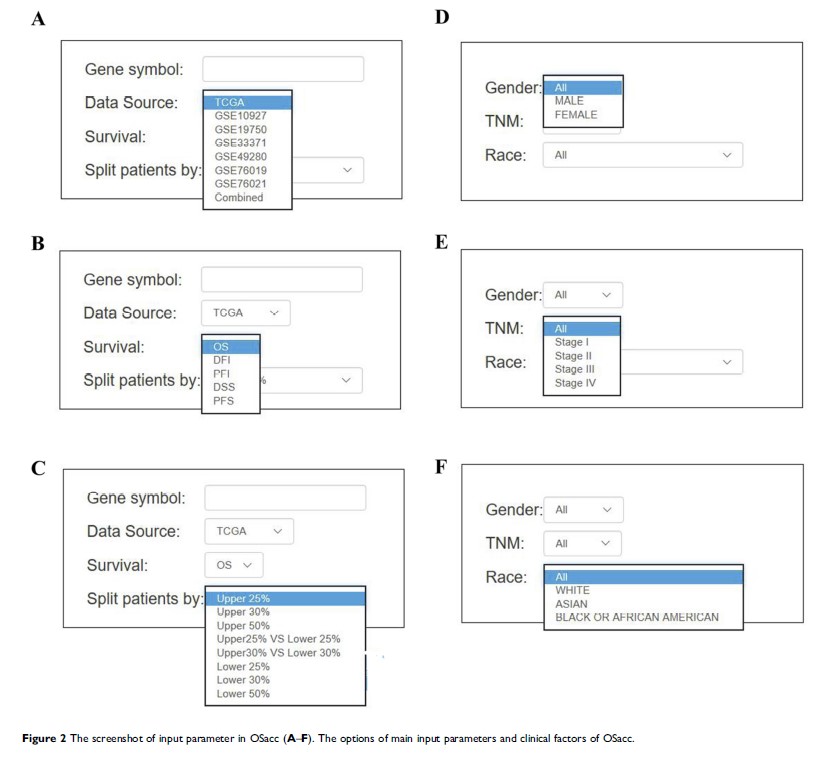
Abstract: Gene expression profiling data with long-term clinical follow-up information are great resources to screen, develop, evaluate and validate prognostic biomarkers in translational cancer research. However, an easy-to-use interactive online tool is needed to analyze these profiling and clinical data. In the current work, we developed OSacc (Online consensus Survival analysis of ACC), a web tool that provides rapid and user-friendly survival analysis based on seven independent transcriptomic profiles with long-term clinical follow-up information of 259 ACC patients gathered from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases. OSacc allows researchers and clinicians to evaluate the prognostic value of genes of interest by Kaplan–Meier (KM) survival plot with hazard ratio (HR) and log-rank test in ACC. OSacc is freely available at http://bioinfo.henu.edu.cn/ACC/ACCList.jsp.
Keywords: ACC, prognostic marker, over survival, web tool, KM plot