111314
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

一个新发现的耐药基因 rfaF 存在于幽门螺杆菌中
Authors Lin J, Zhang X, Wen Y, Chen H, She F
Received 16 September 2019
Accepted for publication 30 October 2019
Published 12 November 2019 Volume 2019:12 Pages 3507—3514
DOI https://doi.org/10.2147/IDR.S231152
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Melinda Thomas
Peer reviewer comments 2
Editor who approved publication: Dr Eric Nulens
Background: The purpose of this study was to understand the function of rfaF gene in Helicobacter pylori antibiotic resistance.
Methods: The gene homologous recombination method was used for knockout and complementation of H. pylori rfaF gene. Various constructed strains were analysed for drug sensitivity to amoxicillin (AMO), tetracycline (TET), clarithromycin (CLA), metronidazole (MET), levofloxacin (LEV), and chloramphenicol (CHL) by agar plate dilution method. Drug sensitivity was further confirmed using a growth inhibition curve. Ethidium bromide (EB) accumulation experiments were performed to assess cell membrane permeability. PCR and sequence analysis were used to detect the rfaF gene.
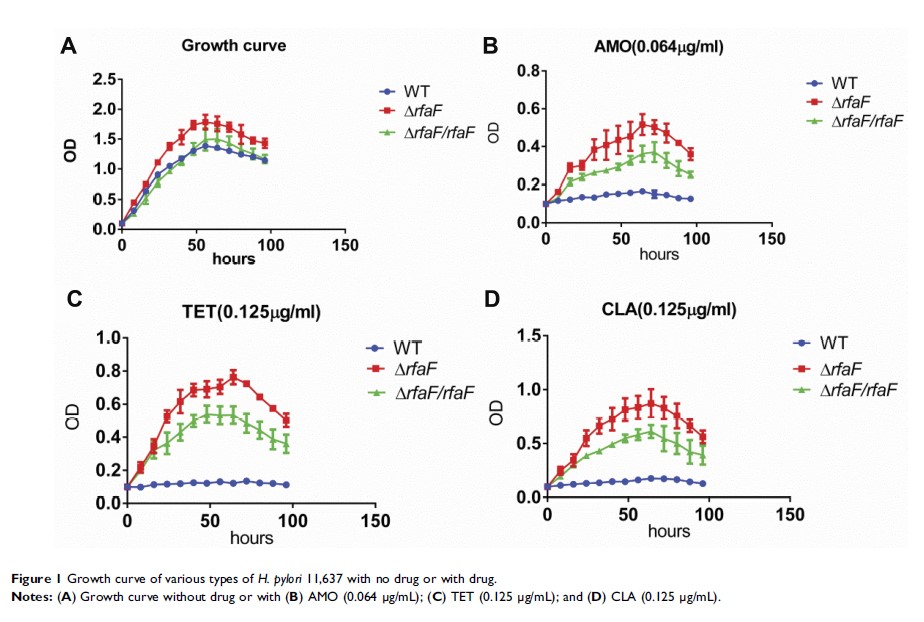
Results: The minimum inhibitory concentrations (MIC) of TET, CHL, AMO, and CLA in 11,637 rfaF knockout strain (ΔrfaF strain) were 4, 4, 2, and 2 times higher than those in 11,637 wild type (WT) strain, respectively. A multidrug-resistant (MDR) ΔrfaF strain also displayed the same trend; however, the degrees of increase were relatively small. Growth inhibition experiments indicated that the growth of the 11,637 ΔrfaF strain was higher with antibiotics at the MIC of the 11,637 WT strain than that of 11,637 rfaF -complemented strain (ΔrfaF/rfaF strain), whereas the 11,637 WT strain did not exhibit any growth. The 11,637 ΔrfaF strain was significantly reduced compared with the cumulative EB fluorescence intensity of the 11,637 WT and of 11,637ΔrfaF/rfaF strain, and the same trend appeared in the MDR strain. Among the 10 clinical strains, 9 clinical strains were found to have mutations in the conserved sequence of rfaF amino acids.
Conclusion: We found a new drug resistance gene, rfaF , in H. pylori , which changes the permeability of cell membrane to confer cross-resistance to AMO, TET, CLA, and CHL and is involved in clinical strain drug resistance. It can be used as a drug target.
Keywords: rfaF , amoxicillin, clarithromycin, tetracycline, resistance