110932
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

杂交基因组装配及使用纳米孔测序和 Illumina 公司的测序技术验证泛耐药肺炎克雷伯菌菌株
Authors Ruan Z, Wu J, Chen H, Draz MS, Xu J, He F
Received 28 November 2019
Accepted for publication 11 January 2020
Published 21 January 2020 Volume 2020:13 Pages 199—206
DOI https://doi.org/10.2147/IDR.S240404
Checked for plagiarism Yes
Review by Single-blind
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Background: The prevalence of multidrug-resistant Klebsiella pneumoniae is increasingly being implicated worldwide in a variety of infections with high mortalities. Here, we report the complete genome sequence of K. pneumoniae strain KP58, a pandrug-resistant K. pneumoniae strain that exhibits high levels of resistance to colistin and tigecycline in China.
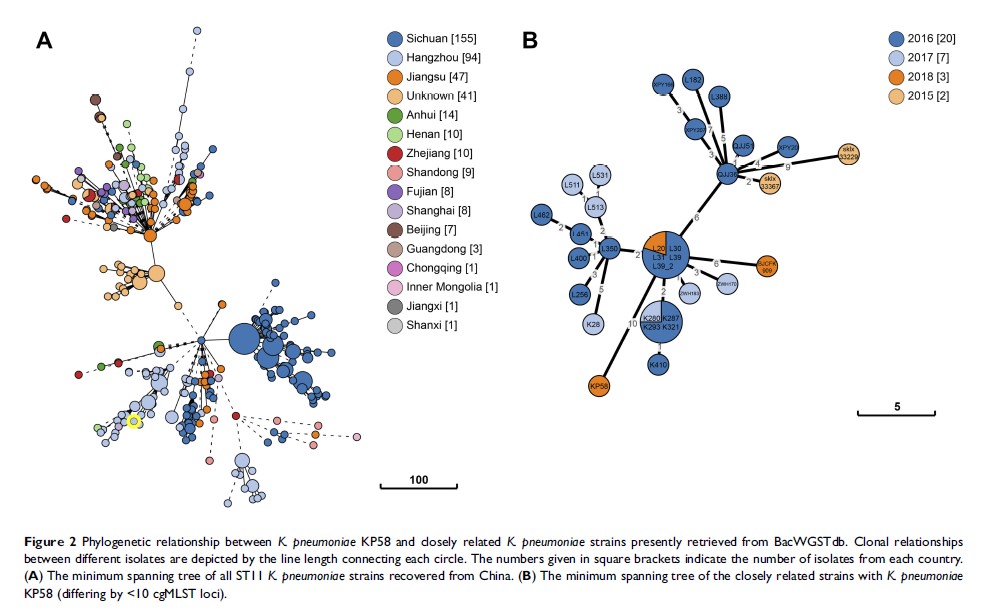
Methods: The K. pneumoniae strain KP58 was recovered from a urine sample of a female patient hospitalized in a tertiary hospital in Hangzhou, China. Antimicrobial susceptibility testing was performed and the minimum inhibitory concentrations (MICs) were determined. Whole-genome sequencing was performed using Illumina and Oxford nanopore sequencing technologies. Genomic features, antimicrobial resistance genes and virulence genes were comprehensively analysed by various bioinformatics approaches. In addition, genomic epidemiological and phylogenetic analyses of K. pneumoniae KP58 and closely related isolates were performed using the core genome multilocus sequence typing (cgMLST) analysis in BacWGSTdb, an online bacterial whole-genome sequence typing and source tracking database.
Results: K. pneumoniae KP58 was resistant to all antimicrobial agents tested, including tigecycline and colistin. Combining the two sequencing technologies allowed a high-quality complete genome sequence of K. pneumoniae KP58 comprising one circular chromosome and five circular plasmids to be obtained. This strain harbours a variety of acquired antimicrobial resistance and virulence determinants. It also carried an ISKpn26 -like insertion in the disrupted mgrB gene, which confers colistin resistance. The tigecycline resistance was associated with overexpression of the AcrAB efflux system. The closest relative of K. pneumoniae KP58 was another clinical isolate recovered from Hangzhou that differed by only 10 cgMLST loci.
Conclusion: The dataset presented in this study provides essential insights into the evolution of antimicrobial-resistant K. pneumoniae in hospital settings and assists in the development of effective control strategies. Appropriate surveillance and control measures are essential to prevent its further dissemination.
Keywords: Klebsiella pneumoniae , whole-genome sequencing, nanopore, hybrid assembly, pandrug resistance