110932
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

人结直肠癌配对肿瘤和邻近的正常组织中肠道微生物群的比较
Authors Sheng QS, He KX, Li JJ, Zhong ZF, Wang FX, Pan LL, Lin JJ
Received 1 June 2019
Accepted for publication 12 December 2019
Published 21 January 2020 Volume 2020:13 Pages 635—646
DOI https://doi.org/10.2147/OTT.S218004
Checked for plagiarism Yes
Review by Single-blind
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Nicola Silvestris
Background: To understand the biological effect of gut microbiome on the progression of colorectal cancer (CRC), we sequenced the V3-V4 region of the 16S rRNA gene to illustrate the overall structure of microbiota in the CRC patients.
Methods: In this study, a total of 66 CRC patients were dichotomized into different groups based on the following characteristics: paired tumor and adjacent normal tissues, distal and proximal CRC segments, MMR (-) and MMR (+), different TNM staging and clinic tumor staging.
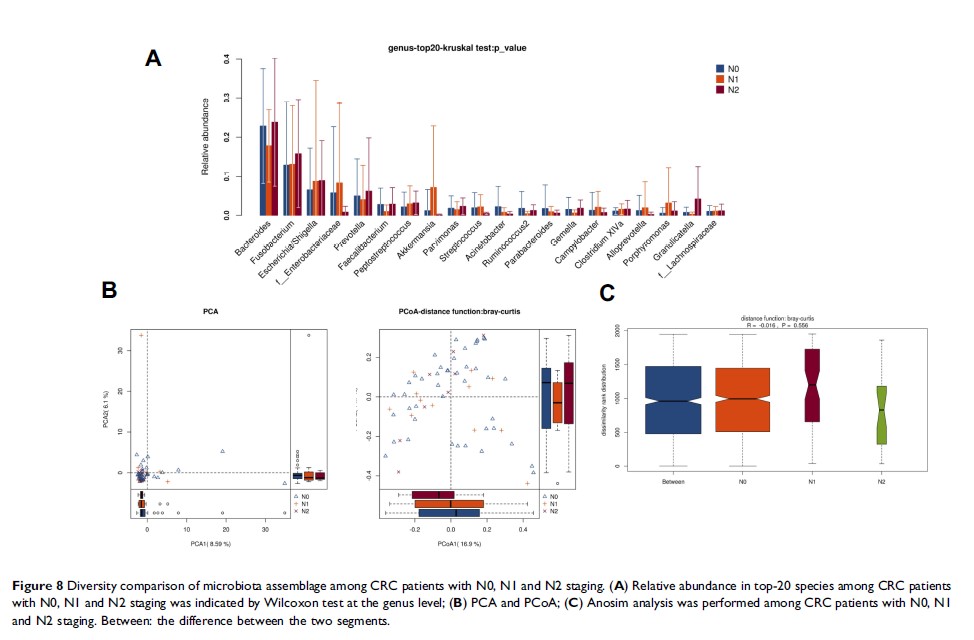
Results: By sequencing and comparing the microbial assemblages, our results indicated that 7 microbe genus (Fusobacterium, Faecalibacterium, Akkermansia, Ruminococcus2, Parabacteroides, Streptococcus , and f_Ruminococcaceae ) were significantly different between tumor and adjacent normal tissues; and 5 microbe genus (Bacteroides, Fusobacterium, Faecalibacterium, Parabacteroides , and Ruminococcus2 ) were significantly different between distal and proximal CRC segments; only 2 microbe genus (f_Enterobacteriaceae and Granulicatella ) were significantly different between MMR (-) and MMR (+); but there was no significant microbial difference were detected neither in the TNM staging nor in the clinic tumor staging.
Conclusion: All these findings implied a better understanding of the alteration in the gut microbiome, which may offer new insight into diagnosing and therapying for CRC patients.
Keywords: colorectal cancer, CRC, gut microbiota, 16S rRNA sequencing, TNM staging, MMR